Page 127 - ITPS-7-2
P. 127
INNOSC Theranostics and
Pharmacological Sciences PI3K-α inhibitors for cancer immunotherapy
properties. Furthermore, bipyridine is capable of inducing protein-ligand complexes by providing additional attractive
chirality through ring functionalization or restricted forces and complementarity between the binding partners.
rotation (atropisomerism), thus increasing its relevance in These suggest that T85 exhibits an adequate number of
asymmetry-based applications. However, the inclusion of interactions, indicating optimal complementarity between
fluorine in the design of T85 is still a limited study, and its the ligand and the receptor.
full potential has yet to be clinically verified. Figure 13 describes the non-covalent interaction of the
Table 4 summarizes the performance comparison hydrophobic type established in the enclosure of T85 by
between the reference hit compound and T85 obtained several hydrophobic amino acid residues. Interestingly, the
from the IPP under the MCS-constrained docking type result revealed that 11 hydrophobic amino acid residues
and 3D-QSAR model prediction. To compare docking enveloped T85, with more isoleucine (ILE) amino acid
scores, a docking score of -9.88 was better than -9.51. This residues involved in the ligand-receptor hydrophobic
meant that T85 was predicted to have a stronger binding interaction network at different positions in the protein
affinity or interaction with the target protein compared to sequence. However, the presence of multiple hydrophobic
Candidate 1. In addition, Table 4 indicates that Candidate 1
possessed a lower biological activity (6.51) than T85 (8.25)
in inhibiting the target protein.
3.6.2. T85-6PYS interactions
In this work, we studied the interactive behavior of T85
within the receptor grid of the target protein in a 2D space.
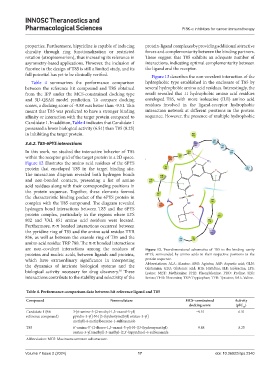
Figure 12 illustrates the amino acid residues of the 6PYS
protein that enveloped T85 in the target binding site.
The interaction diagram revealed both hydrogen bonds
and non-bonded contacts, presenting a list of amino
acid residues along with their corresponding positions in
the protein sequence. Together, these elements formed
the characteristic binding pocket of the 6PYS protein in
complex with the T85 compound. The diagram revealed
hydrogen bond interactions between T85 and the 6PYS
protein complex, particularly in the regions where LYS
802 and VAL 851 amino acid residues were located.
Furthermore, π-π bonded interactions occurred between
the pyridine ring of T85 and the amino acid residue TYR
836, as well as between the oxazole ring of T85 and the
amino acid residue TRP 780. The π-π bonded interactions
are non-covalent interactions among the residues of Figure 12. Two-dimensional schematics of T85 in the binding cavity
proteins and nucleic acids, between ligands and proteins, 6PYS, surrounded by amino acids in their respective positions in the
which have extraordinary significance in interpreting protein sequence.
the dynamics of intricate biological systems and the Abbreviations: ALA: Alanine: ARG: Aginine; ASP: Aspartic acid; GLN:
Glutamine; GLU: Glutamic acid; HIS: Histidine; ILE: Isoleucine; LYS:
92
biological activity necessary for drug discovery. These Lysine; MET: Methionine; PHE: Phenylalanine; PRO: Proline; SER:
interactions contribute to the stability and selectivity of the Serine; THR: Threonine; TRP: Tryptophan; TYR: Tyrosine; VAL: Valine.
Table 4. Performance comparison data between hit reference ligand and T85
Compound Nomenclature MCS‑ constrained Activity
docking score (pIC )
50
Candidate 1 (hit 3-[6-amino-5-(2-methyl-1,3-oxazol-5-yl) −9.51 6.51
reference compound) pyridin-3-yl]-N-{[3-(hydroxymethyl) oxetan-3-yl]
methyl}-4-methylbenzene-1-sulfonamide
T85 6’-amino-5’-(2-fluoro-1,3-oxazol-5-yl)-N-{[3-(hydroxymethyl) −9.88 8.25
oxetan-3-yl] methyl}-3-methyl-[2,3’-bipyridine]-6-sulfonamide
Abbreviation: MCS: Maximum common substructure.
Volume 7 Issue 2 (2024) 17 doi: 10.36922/itps.2340