Page 13 - MI-2-4
P. 13
Microbes & Immunity Regulation of Staphylococcus aureus CP biosynthesis
Figure 2. Amino acid (a.a.) homology comparison of enzymes involved in the biosynthesis of CP5 and CP8 in Staphylococcus aureus. A total of 16 enzymes
encoded by the cap operon were compared. The approximate number of a.a. for each protein is indicated on the top scale. The percentage of a.a. identity
between CP5 and CP8 is displayed. The comparison revealed that 12 out of 16 enzymes are highly conserved between CP5 and CP8, sharing more than
97% of amino acid identity. The remaining enzymes (CapH, CapI, CapJ, and CapK) exhibit <43% homology. Figure created using PowerPoint software.
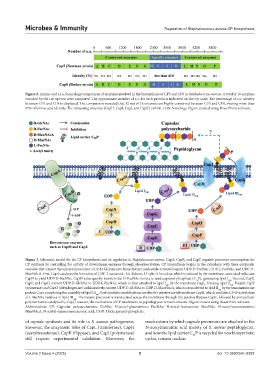
Figure 3. Schematic model for the CP biosynthesis and its regulation in Staphylococcus aureus. CapA, CapB, and CapC regulate precursor consumption for
CP synthesis by controlling the activity of downstream enzymes through phosphorylation. CP biosynthesis begins in the cytoplasm with three enzymatic
cascades that convert the universal precursor UDP-D-GlcNAc into three distinct nucleotide-activated sugars: UDP-D-FucNAc, UDP-L-FucNAc, and UDP-D-
ManNAcA. First, CapD catalyzes the formation of UDP-2-acetamido-2,6-dideoxy-D-xylo−4-hexulose, which is reduced by the membrane-associated reductase
CapN to yield UDP-D-FucNAc. CapM subsequently transfers the D-FucNAc moiety to undecaprenyl-phosphate (C P), generating lipid I . Second, CapE,
cap
55
CapF, and CapG convert UDP-D-GlcNAc to UDP-L-FucNAc, which is then attached to lipid I by the transferase CapL, forming lipid II . Finally, CapP
cap
cap
(epimerase) and CapO (dehydrogenase) collaboratively convert UDP-D-GlcNAc to UDP-D-ManNAcA, which is transferred to lipid II by the transmembrane
cap
protein CapI, completing the assembly of lipid III . Post-synthetic modifications involve the putative acetyltransferase CapH, which mediates C3-O-acetylation
cap
of L-FucNAc residues in lipid III . The mature precursor is translocated across the membrane through the putative flippase CapK, followed by extracellular
cap
polymerization catalyzed by CapJ. However, the mechanism of CP attachment to peptidoglycan remains obscure. Figure created using PowerPoint software.
Abbreviations: CP: Capsular polysaccharides; GlcNAc: N-acetyl-glucosamine; FucNAc: N-acetyl-fucosamine; ManNAc: N-acetyl-mannosamine;
ManNAcA: N-acetyl-mannosaminuronic acid; C55P: Undecaprenyl-phosphate.
of capsule synthesis and its role in S. aureus pathogenesis. mechanisms by which capsule precursors are attached to the
However, the enzymatic roles of CapL (transferase), CapH N-acetylmuramic acid moiety of S. aureus peptidoglycan,
(acetyltransferase), CapK (flippase), and CapJ (polymerase) and how the lipid carrier C P is recycled for new biosynthetic
55
still require experimental validation. Moreover, the cycles, remain unclear.
Volume 2 Issue 4 (2025) 5 doi: 10.36922/mi.8392