Page 53 - TD-3-3
P. 53
Tumor Discovery Somatic mutations in POLE and POLD1 in colorectal cancer
variants were non-synonymous variants, which resulted No recurrent mutations or double heterozygous mutations
in amino acid substitutions in the endonuclease domains were identified among the new POLE/POLD1 alterations.
of POLE and POLD1. One mutation in the POLE resulted Furthermore, the previously described germline POLE
in a stop codon, which is predicted to result in pol ɛ L424D or POLD1 S478N mutations were not detected in our
22
translation termination. With the exception of the POLE cohort. In our previous study, we performed NGS analysis
D414N variants, all the other variants in POLE and POLD1 on the same cohort of samples that was used here. Based on
were found in the primary tumors (Table 2). However, no our findings, only four out of ten samples with mutations
variant was found in the corresponding normal tissues of in the POLE and POLD1 genes exhibited a hypermutated
these cases, thereby confirming that these novel alterations phenotype. These samples were more likely to have a full
were somatic mutations rather than germline SNPs. Sanger complement of mutations in APC, KRAS, SMAD4, and
sequencing data also validated the novel alterations as TP53. In contrast, the remaining six out of ten samples were
somatic and not germline (Figures 1A-F). more likely to have subclonal mutations (Table 2).
A B
C D
E F
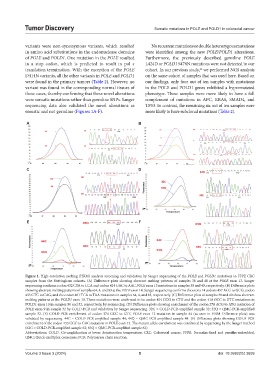
Figure 1. High-resolution melting (HRM) analysis screening and validation by Sanger sequencing of the POLE and POLD1 mutations in FFPE CRC
samples from the Nottingham cohorts. (A) Difference plots showing aberrant melting patterns of samples 38 and 40 at the POLE exon 13. Sanger
sequencing confirms codon 432 CTA to CCA and codon 414 GAC to AAC POLE exon 13 mutations in samples 38 and 40, respectively. (B) Difference plots
showing aberrant melting patterns of samples 64, 4, and 83 at the POLE exon 14. Sanger sequencing confirms the exons 14 codons 457 ACG to GCG, codon
455 CTG to CAG, and the codon 461 TCA to TAA mutations in samples 64, 4, and 83, respectively. (C) Difference plots of samples 38 and 40 show aberrant
melting patterns at the POLD1 exon 10. These mutations were confirmed to be codon 404 CCG to CTG and the codon 410 GCC to GTC mutations in
POLD1 exon 10 in samples 50 and 52, respectively, by sequencing. (D) Difference plots showing enrichment of the codon 278 ACG to ATG mutation of
POLE exon 9 in sample 32 by COLD-PCR and validation by Sanger sequencing. 32C = COLD-PCR-amplified sample 32; 32Q = QMC-PCR-amplified
sample 32. (E) COLD-PCR enrichment of codon 374 GCC to GTC POLE exon 12 mutation in sample 44 (as seen in HRM Difference plots) was
validated by sequencing. 44C = COLD-PCR-amplified sample 44; 44Q = QMC PCR-amplified sample 44. (F) Difference plots showing COLD-PCR
enrichment of the codon 433 GGC to GAC mutation in POLE exon 13. The mutant allele enrichment was confirmed by sequencing by the Sanger method
(63C = COLD-PCR-amplified sample 63; 63Q = QMC-PCR-amplified sample 63)
Abbreviations: COLD: Co-amplification at lower denaturation temperature; CRC: Colorectal cancer; FFPE: Formalin-fixed and paraffin-embedded;
QMC: Quick-multiplex consensus; PCR: Polymerase chain reaction.
Volume 3 Issue 3 (2024) 5 doi: 10.36922/td.3659