Page 55 - TD-3-3
P. 55
Tumor Discovery Somatic mutations in POLE and POLD1 in colorectal cancer
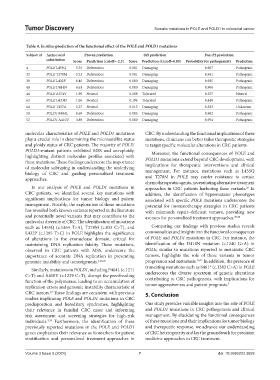
Table 4. In silico prediction of the functional effect of the POLE and POLD1 mutations
Subject id Amino acid Proven prediction Sift prediction Pon‑P2 prediction
substitution Score Prediction (cutoff=‑2.5) Score Prediction (Cutoff=0.05) Probability for pathogenicity Prediction
4 POLE L455Q −5.36 Deleterious 0.001 Damaging 0.907 Pathogenic
32 POLE T278M −5.23 Deleterious 0.001 Damaging 0.941 Pathogenic
38 POLE L432P −6.46 Deleterious 0.000 Damaging 0.925 Pathogenic
40 POLE D414N −4.61 Deleterious 0.000 Damaging 0.906 Pathogenic
44 POLE A374V −1.95 Neutral 0.088 Tolerated 0.257 Neutral
63 POLE G433D −1.06 Neutral 0.194 Tolerated 0.848 Pathogenic
64 POLE T457A −2.27 Neutral 0.013 Damaging 0.523 Unknown
50 POLD1 P404L −8.65 Deleterious 0.005 Damaging 0.962 Pathogenic
52 POLD1 A410V −3.89 Deleterious 0.000 Damaging 0.954 Pathogenic
molecular characteristics of POLE and POLD1 mutations CRC. By understanding the functional implications of these
play a crucial role in determining the microsatellite status mutations, clinicians can better tailor therapeutic strategies
and ploidy status of CRC patients. The majority of POLE/ to target specific molecular alterations in CRC patients.
POLD1-mutant patients exhibited MSS and aneuploidy, Moreover, the functional consequences of POLE and
highlighting distinct molecular profiles associated with
these mutations. These findings underscore the importance POLD1 mutations extend beyond CRC development, with
implications for therapeutic interventions and clinical
of molecular subtyping in understanding the underlying management. For instance, mutations such as L455Q
biology of CRC and guiding personalized treatment
approaches. and T278M in POLE may confer resistance to certain
chemotherapeutic agents, necessitating alternative treatment
In our analysis of POLE and POLD1 mutations in approaches in CRC patients harboring these variants. In
25
CRC patients, we identified several key mutations with addition, the identification of hypermutator phenotypes
significant implications for tumor biology and patient associated with specific POLE mutations underscores the
management. Notably, the exploration of these mutations potential for immunotherapy strategies in CRC patients
has revealed both known variants reported in the literature with mismatch repair–deficient tumors, providing new
and potentially novel variants that may contribute to the avenues for personalized treatment approaches. 29,30
molecular diversity of CRC. The identification of mutations
such as L455Q (c.1364 T>A), T278M (c.833 C>T), and Comparing our findings with previous studies reveals
L432P (c.1295 T>C) in POLE highlights the significance commonalities and insights into the functional consequences
of alterations in the exonuclease domain, critical for of POLE and POLD1 mutations in CRC. For instance, the
maintaining DNA replication fidelity. These mutations, identification of the D414N mutation (c.1240 G>A) in
observed in CRC patients with MSS, underscore the POLE, similar to mutations reported in metastatic CRC
importance of accurate DNA replication in preventing tumors, highlights the role of these variants in tumor
genomic instability and tumorigenesis. 1,25,26 progression and metastasis. 26,31 In addition, the presence of
truncating mutations such as S461* (c.1382 C>A) in POLE
Similarly, mutations in POLD1, including P404L (c.1211
C>T) and A410V (c.1229 C>T), disrupt the proofreading underscores the diverse spectrum of genetic alterations
function of the polymerase, leading to an accumulation of contributing to CRC pathogenesis, with implications for
3
replication errors and genomic instability characteristic of tumor aggressiveness and patient prognosis.
27
CRC tumors. These findings are consistent with previous 5. Conclusion
studies implicating POLE and POLD1 mutations in CRC
predisposition and hereditary syndromes, highlighting Our study provides valuable insights into the role of POLE
their relevance in familial CRC cases and informing and POLD1 mutations in CRC pathogenesis and clinical
risk assessment and screening strategies for high-risk management. By elucidating the functional consequences
individuals. Furthermore, the identification of these of these mutations and their implications for tumor biology
3,28
previously reported mutations in the POLE and POLD1 and therapeutic response, we advance our understanding
genes emphasizes their relevance as biomarkers for patient of CRC heterogeneity and lay the groundwork for precision
stratification and personalized treatment approaches in medicine approaches in CRC treatment.
Volume 3 Issue 3 (2024) 7 doi: 10.36922/td.3659