Page 63 - TD-4-1
P. 63
Tumor Discovery Drug repurposing for pancreatic cancer via AI
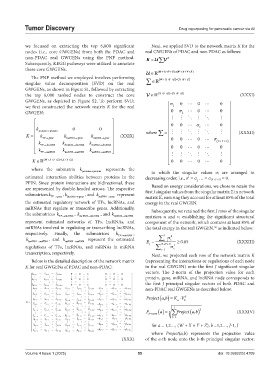
we focused on extracting the top 6,000 significant Next, we applied SVD to the network matrix K for the
nodes (i.e., core GWGENs) from both the PDAC and real GWGENs of PDAC and non-PDAC as follows:
non-PDAC real GWGENs using the PNP method. K U V T
Subsequently, KEGG pathways were utilized to annotate
these core GWGENs. (WX )YZ (WX )YZ
The PNP method we employed involves performing WX YZ
XY Z
singular value decomposition (SVD) on the real
GWGENs, as shown in Figure S1, followed by extracting
)Z
the top 6,000 ranked nodes to construct the core (XY (X )YZ (XXXI)
GWGENs, as depicted in Figure S2. To perform SVD, σ 0 0 0
we first constructed the network matrix K for the real 1
GWGEN: σ 2 0 0 0
0 0 σ i 0
k protein protein 0 0 where = (XXXII)
K k k k (XXIX) ∑
TF gene lncRNA gene miRNA gene 0 0 σ ( ++Z ) 0
XY
k TF lncRRNA k lncRNA lncRNA k miRNA lncRNA
k k k 0 0 0 0
TF miRNA lncRNA miRNA miRNAA miRNA
( WX YZ ) ( XY Z )
K 0 0 0 0
where the submatrix k protein↔ protein represents the in which the singular values σ I are arranged in
estimated interaction abilities between proteins in the decreasing order, i.e., σ ≥ σ … ≥ σ (X+Y+Z) ≥ 0.
1
PPIN. Since protein interactions are bidirectional, these 2
are represented by double-headed arrows. The respective Based on energy considerations, we chose to retain the
submatrices k TF→ gene , k lncRNA→ gene , and k miRNA→ gene represent first J singular values from the singular matrix Σ in network
matrix K, ensuring they account for at least 85% of the total
the estimated regulatory network of TFs, lncRNAs, and energy in the real GWGEN.
miRNAs that regulate or transcribe genes. Additionally, Subsequently, we retained the first J rows of the singular
the submatrices k TF→ lncRNA , k lncRNA→ lncRNA , and k miRNA→ lncRNA matrices u and v, establishing the significant structural
represent estimated networks of TFs, lncRNAs, and component of the network, which contains at least 85% of
miRNAs involved in regulating or transcribing lncRNAs, the total energy in the real GWGEN, as indicated below:
30
respectively. Finally, the submatrices k TF→ miRNA , J
k lncRNA→ miRNA , and k miRNA→ miRNA represent the estimated E i1 2 i 085. (XXXIII)
regulations of TFs, lncRNAs, and miRNAs in miRNA j XY Z c 2
c1
transcription, respectively. Next, we projected each row of the network matrix K
Below is the detailed description of the network matrix (representing the interactions or regulations of each node
K for real GWGENs of PDAC and non-PDAC: in the real GWGEN) onto the first J significant singular
vectors. The 2-norm of the projection value for each
protein, gene, miRNA, and lncRNA node corresponds to
the first J principal singular vectors of both PDAC and
non-PDAC real GWGENs as described below:
Projecta b, K V b T
a
J 2
,
P 2 norm a 2 Projecta b (XXXIV)
b 1
for a = 1,2…, (W + X + Y + Z), b =1,2…, J-1, J
where Project(a,b) represents the projection value
(XXX) of the a-th node onto the b-th principal singular vector;
Volume 4 Issue 1 (2025) 55 doi: 10.36922/td.4709