Page 104 - AJWEP-v22i3
P. 104
Sonsare, et al.
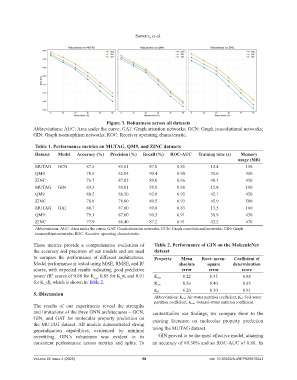
Figure 3. Robustness across all datasets
Abbreviations: AUC: Area under the curve; GAT: Graph attention networks; GCN: Graph convolutional networks;
GIN: Graph isomorphism networks; ROC: Receiver operating characteristic.
Table 1. Performance metrics on MUTAG, QM9, and ZINC datasets
Dataset Model Accuracy (%) Precision (%) Recall (%) ROC‑AUC Training time (s) Memory
usage (MB)
MUTAG GCN 87.5 85.01 87.0 0.85 12.4 150
QM9 78.3 85.04 90.4 0.90 35.6 410
ZINC 76.7 87.05 89.0 0.86 40.3 450
MUTAG GIN 89.3 88.01 88.0 0.88 15.8 180
QM9 80.5 88.20 92.8 0.92 42.1 450
ZINC 78.8 78.80 89.5 0.93 45.9 500
MUTAG GAT 88.7 87.00 89.0 0.83 13.5 160
QM9 79.1 87.00 90.5 0.91 38.9 430
ZINC 77.9 86.40 87.2 0.91 42.2 470
Abbreviations: AUC: Area under the curve; GAT: Graph attention networks; GCN: Graph convolutional networks; GIN: Graph
isomorphism networks; ROC: Receiver operating characteristic.
These metrics provide a comprehensive evaluation of Table 2. Performance of GIN on the MoleculeNet
the accuracy and precision of our models and are used dataset
to compare the performance of different architectures. Property Mean Root‑ mean‑ Coefficient of
Model performance is tested using MAE, RMSE, and R absolute square determination
2
scores, with expected results indicating good predictive error error score
power (R scores of 0.88 for K , 0.85 for K w, and 0.91 K ow 0.22 0.35 0.88
2
ow
a
for K_d), which is shown in Table 2.
K aw 0.26 0.40 0.85
5. Discussion K d 0.20 0.30 0.91
Abbreviations: K aw: Air-water partition coefficient; K d: Soil-water
partition coefficient; K ow: Octanol-water partition coefficient.
The results of our experiments reveal the strengths
and limitations of the three GNN architectures – GCN, contextualize our findings, we compare them to the
GIN, and GAT for molecular property prediction on existing literature on molecular property prediction
the MUTAG dataset. All models demonstrated strong
generalization capabilities, evidenced by minimal using the MUTAG dataset.
overfitting. GIN’s robustness was evident in its GIN proved to be the most effective model, attaining
consistent performance across metrics and splits. To an accuracy of 89.30% and an ROC-AUC of 0.88. Its
Volume 22 Issue 3 (2025) 98 doi: 10.36922/AJWEP025070041