Page 10 - EJMO-9-1
P. 10
Eurasian Journal of Medicine and
Oncology
Single-cell RNA-seq in malignant skin tumors
common types of skin cancer are basal cell carcinoma the MAPK pathway and inducing alterations in the cell
(BCC), squamous cell carcinoma (SCC), and cutaneous cycle. Despite these advancements, bulk sequencing
7
melanoma (CM). Although BCC and SCC have relatively analyses conducted on overall tissue often overlook
3
low metastatic potential, a subset of these tumors can lead detailed information about the cellular background.
4
to widespread complications through local infiltration and Averaged molecular results derived from mixed
tissue destruction. In contrast, CM has a higher potential for tissues may obscure the crucial role played by rare cell
malignant metastasis, making it the deadliest form of skin subpopulations in the disease progression. Furthermore,
cancer. Commonly employed treatment methods included understanding the intercellular communication
surgical resection, radiation therapy, and chemotherapy. mechanisms that driving tumor infiltration and malignant
Recently, the emergence of immune checkpoint inhibitors transformation within tissues remains challenging with
has introduced promising new treatment modalities that bulk sequencing methodologies.
have demonstrated significant effectiveness in treating skin
cancer. Research suggests that the malignancy, prognosis, 2. Introduction to single-cell sequencing
and treatment effectiveness of skin cancer are closely technology
linked to the molecular characteristics present within the In recent years, single-cell sequencing, particularly single-
tumors. 5 cell RNA-seq (scRNA-seq), has gained significant traction
High-throughput sequencing technologies offer and widespread application. 8-11 Comparatively, another
robust tools for the thorough characterization of high-throughput sequencing technology, single nucleus
molecular features within tumors. Sequencing methods RNA-seq, only extracts RNA from the nucleus of individual
such as DNA sequencing, RNA sequencing (RNA-seq), cells. This limitation may result in the loss of important
whole-genome bisulfite sequencing, and chromatin transcripts, particularly mature mRNA that has already
immunoprecipitation sequencing have facilitated undergone translation. In contrast, scRNA-seq technology
detailed descriptions of tumor cells from various provides comprehensive information on gene expression
perspectives encompassing the genome, transcriptome, and genomic information at the single-cell level, capturing
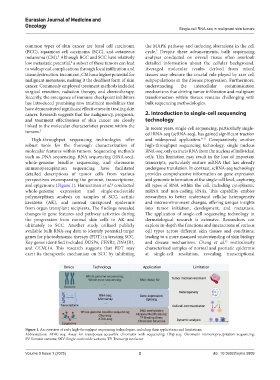
and epigenome (Figure 1). Hameetman et al. conducted all types of RNA within the cell, including cytoplasmic
6
whole-genome expression and single-nucleotide mRNA and non-coding RNAs. This capability enables
polymorphism analysis on samples of SCC, actinic researchers to better understand cellular heterogeneity
keratosis (AK), and normal unexposed epidermis and microenvironment changes, offering unique insights
from organ transplant recipients. The findings revealed into tumor initiation, development, and metastasis.
changes in gene features and pathway activities during The application of single-cell sequencing technology in
the progression from normal skin cells to AK and dermatological research is extensive. Researchers can
ultimately to SCC. Another study utilized publicly explore in-depth the functions and interactions of various
available bulk RNA-seq data to identify potential target cell types across different skin tissues and conditions,
genes for photodynamic therapy (PDT) in treating SCC. leading to a more nuanced understanding of skin biology
Key genes identified included DUSP6, EFNB2, DNAJB1, and disease mechanisms. Cheng et al. meticulously
12
and CCNL14. This research suggests that PDT may characterized samples of normal and psoriatic epidermis
exert its therapeutic mechanism on SCC by inhibiting at single-cell resolution, revealing transcriptional
Figure 1. An overview of early high-throughput sequencing technologies, including their applications and limitations.
Abbreviations: ATAC-seq: Assay for transposase-accessible chromatin with sequencing; Chip-seq: Chromatin immunoprecipitation sequencing;
SV: Somatic variants; SNV: Single-nucleotide variants; TF: Transcription factor.
Volume 9 Issue 1 (2025) 2 doi: 10.36922/ejmo.5809