Page 282 - EJMO-9-2
P. 282
Eurasian Journal of
Medicine and Oncology QGJSF multi-target mechanisms in osteoporosis
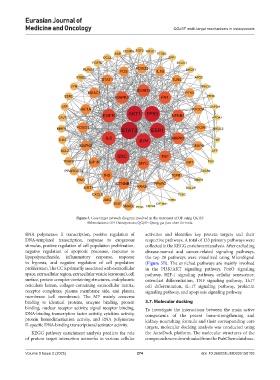
Figure 3. Core target network diagram involved in the treatment of OP using QGJSF
Abbreviations: OP: Osteoporosis; QGJSF: Qiang-gu-jian-shen formula.
RNA polymerase II transcription, positive regulation of activities and identifies key protein targets and their
DNA-templated transcription, response to exogenous respective pathways. A total of 133 primary pathways were
stimulus, positive regulation of cell population proliferation, collected in the KEGG enrichment analysis. After excluding
negative regulation of apoptotic processes, response to disease-named and cancer-related signaling pathways,
lipopolysaccharide, inflammatory response, response the top 20 pathways were visualized using MicroSignal
to hypoxia, and negative regulation of cell population (Figure 5B). The enriched pathways are mainly involved
proliferation. The CC is primarily associated with extracellular in the PI3K/AKT signaling pathway, FoxO signaling
space, extracellular region, extracellular vesicle (exosome), cell pathway, HIF-1 signaling pathway, cellular senescence,
surface, protein complex-containing structures, endoplasmic osteoclast differentiation, TNF signaling pathway, Th17
reticulum lumen, collagen-containing extracellular matrix, cell differentiation, IL-17 signaling pathway, prolactin
receptor complexes, plasma membrane side, and plasma signaling pathway, and apoptosis signaling pathway.
membrane (cell membrane). The MF mainly concerns
binding to identical proteins, enzyme binding, protein 3.7. Molecular docking
binding, nuclear receptor activity, signal receptor binding, To investigate the interactions between the main active
DNA-binding transcription factor activity, cytokine activity, components of the potent bone-strengthening and
protein homodimerization activity, and RNA polymerase kidney-nourishing formula and their corresponding core
II-specific DNA-binding transcriptional activator activity. targets, molecular docking analysis was conducted using
KEGG pathway enrichment analysis predicts the role the AutoDock platform. The molecular structures of the
of protein target interaction networks in various cellular compounds were downloaded from the PubChem database,
Volume 9 Issue 2 (2025) 274 doi: 10.36922/EJMO025150103