Page 285 - EJMO-9-2
P. 285
Eurasian Journal of
Medicine and Oncology QGJSF multi-target mechanisms in osteoporosis
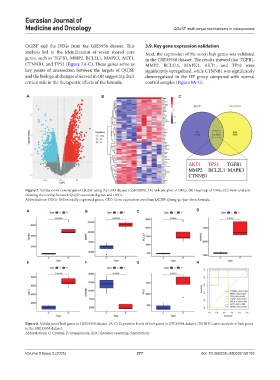
QGJSF and the DEGs from the GSE5958 dataset. This 3.9. Key gene expression validation
analysis led to the identification of seven shared core Next, the expression of the seven hub genes was validated
genes, such as TGFB1, MMP2, BCL2L1, MAPK3, AKT1, in the GSE35958 dataset. The results showed that TGFB1,
CTNNB1, and TP53 (Figure 7A-C). These genes serve as MMP2, BCL2L1, MAPK3, AKT1, and TP53 were
key points of intersection between the targets of QGJSF significantly upregulated, while CTNNB1 was significantly
and the biological changes observed in OP, suggesting their downregulated in the OP group compared with normal
critical role in the therapeutic effects of the formula. control samples (Figure 8A-G).
A B C
Figure 7. Validation of core targets of QGJSF using the GEO dataset (GSE35958). (A) Volcano plot of DEGs. (B) Heatmap of DEGs. (C) Venn analysis
showing the overlap between QGJSF-associated genes and DEGs.
Abbreviations: DEGs: Differentially expressed genes; GEO: Gene expression omnibus; QGJSF: Qiang-gu-jian-shen formula.
A B C D
E F G H
Figure 8. Validation of hub genes in GSE35958 dataset. (A-G) Expression levels of hub genes in GSE35958 dataset. (H) ROC curve analysis of hub genes
in the GSE35958 dataset.
Abbreviations: C: Control; P: Osteoporosis; ROC: Receiver operating characteristic.
Volume 9 Issue 2 (2025) 277 doi: 10.36922/EJMO025150103