Page 99 - EJMO-9-3
P. 99
Eurasian Journal of
Medicine and Oncology Novel senescence-based melanoma risk model
A B
C D
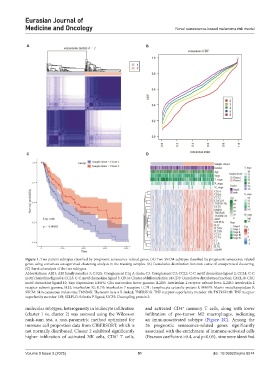
Figure 1. Two patient subtypes classified by prognostic senescence-related genes. (A) Two SKCM subtypes classified by prognostic senescence-related
genes using consensus unsupervised clustering analysis in the training samples. (B) Cumulative distribution function curve of unsupervised clustering.
(C) Survival analysis of the two subtypes.
Abbreviations: ABI3: ABI family member 3; C1QA: Complement C1q A chain; C3: Complement C3; CCL2: C-C motif chemokine ligand 2; CCL4: C-C
motif chemokine ligand 4; CCL5: C-C motif chemokine ligand 5; CD14: Cluster of differentiation 14; CDF: Cumulative distribution function; CXCL10: CXC
motif chemokine ligand 10; Exp: Expression; GMFG: Glia maturation factor gamma; IL2RB: Interleukin 2 receptor subunit beta; IL2RG: Interleukin 2
receptor subunit gamma; IL32: Interleukin 32; IL7R: Interleukin 7 receptor; LCP1: Lymphocyte cytosolic protein 1; MMP9: Matrix metallopeptidase 9;
SKCM: Skin cutaneous melanoma; TMSB4X: Thymosin beta 4 X-linked; TNFRSF1B: TNF receptor superfamily member 1B; TNFRSF13B: TNF receptor
superfamily member 13B; SELPLG: Selectin P ligand; UCP2: Uncoupling protein 2.
molecular subtypes, heterogeneity in leukocyte infiltration and activated CD4 memory T cells, along with lower
+
(cluster 1 vs. cluster 2) was assessed using the Wilcoxon infiltration of pro-tumor M2 macrophages, indicating
rank-sum test, a non-parametric method optimized for an immunoactivated subtype (Figure 2C). Among the
immune cell proportion data from CIBERSORT, which is 26 prognostic senescence-related genes significantly
not normally distributed. Cluster 2 exhibited significantly associated with the enrichment of immune-activated cells
higher infiltration of activated NK cells, CD8 T cells, (Pearson coefficient >0.4, and p<0.05), nine were identified
+
Volume 9 Issue 3 (2025) 91 doi: 10.36922/ejmo.8574