Page 59 - GPD-2-4
P. 59
Gene & Protein in Disease K fragment for resistance gene hunting
A B
Figure 2. The construction of plasmid K fragment vector. (A) Digestion of MCS-free pUC19 (pUC19 -MCS ) and pUC19 vectors with RsaI. Lane 1: As a result
of the digestion of pUC19 vector with RsaI, three bands were formed, corresponding to fragment sizes of 1769 bp, 676 bp and 241 bp. Lane 2: Following
the digestion of pUC19 -MCS vector with RsaI, two bands were formed, corresponding to fragment sizes of 2010 bp and 676 bp. M: Lambda-PstI marker.
(B) Removal of MCS from pUC19 vector. MCS was excised from pUC19 vector by cleaving with EcoRI and HindIII enzymes and the vector was then self-
ligated. The excision of MCS from the vector reduces the number of RsaI restriction sites from 3 to 2 in the vector.
A B C
Figure 3. Insertion of Prom-RBS sequence and confirmation with polymerase chain reaction (PCR). (A) The workflow of insertion to pUC19 -MCS through
the site-directed mutagenesis method. (B) Addition of Prom-RBS into pUC19 -MCS . Lane 1: PCR amplicon (2704 bp) obtained using pUC19 -MCS as DNA
source and P1-pUCPR primer pair. Lane 2: PCR amplicon (2720 bp) obtained using amplicon from Lane 1 as DNA source and P2-pUCPR primer pair.
Lane 3: PCR amplicon (2735 bp) obtained using amplicon from Lane 2 as DNA source and P3-pUCPR primer pair. M: Lambda-PstI marker. (C) PCR
products amplified from plasmid K fragment (pKF) and pUC19 -MCS vectors using prmtrSeq-KsekF primer pair. Lane 1: PCR amplicon was amplified from
pKF and its size was 1079 bp. Lane 2: PCR amplicon was amplified from pUC19 -MCS and its size was 1031 bp.
primers, and pUC19 -MCS was used as a control. The primers A
used amplify regions that contain Prom-RBS. Thus, if pKF
was DNA source, the amplicon size was 1079 bp, but if
the Prom-RBS sequence was not inserted into pUC19 -MCS ,
the amplicon size was 1031 bp (Figure 3C). Furthermore,
the integration of Prom-RBS into the vector was validated B
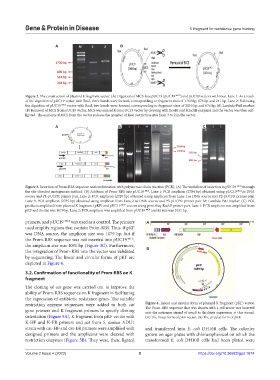
by sequencing. The linear and circular forms of pKF are
depicted in Figure 4.
3.2. Confirmation of functionality of Prom-RBS on K
fragment
The cloning of cat gene was carried out to improve the
ability of Prom-RBS sequence on K fragment in facilitating
the expression of antibiotic resistance genes. The suitable
restriction enzyme sequences were added to both cat Figure 4. Linear and circular form of plasmid K fragment (pKF) vector.
gene primers and K fragment primers to specify cloning The Prom-RBS sequence that was shown with a red arrow was inserted
into the antisense strand of ampR to facilitate expression at this strand.
orientation (Figure 5A). K fragment from pKF vector with (A) The linear form of pKF vector. (B) The circular form of pKF.
K-HF and K-ER primers and cat from S. aureus ADU1
strain with cat-HF and cat-ER primers were amplified with and transferred into E. coli DH10B cells. The colonies
designed primers and the amplicons were cleaved with grown on agar plates with chloramphenicol on which the
restriction enzymes (Figure 5B). They were, then, ligated transformed E. coli DH10B cells had been plated were
Volume 2 Issue 4 (2023) 5 https://doi.org/10.36922/gpd.1674