Page 125 - GPD-3-2
P. 125
Gene & Protein in Disease Bioinformatics to identify gene signatures of CF
1. Introduction signatures of CF using transcriptome profiling of human
bronchial epithelial cells. In this study, we investigated
Each year, thousands of new cystic fibrosis (CF) diagnosis in the specific differentially expressed genes (DEGs), gene
infants within one year of life are reported, and approximately networks, pathways, and the interactions between proteins
1
162,428 individuals are living with CF worldwide. In the associated with CF. We utilized an integrative systems
2
USA, the leading cause of mortality for Caucasians is CF, biology approach to identify DEGs in bronchial epithelial
3
with an incidence rate of one in approximately 15,000–32,000 cells using data from the GEO database.
individuals within this specific ethnic group. According
1
to the European Cystic Fibrosis Society patient registry Combining different fields of study such as computer
and analysis, it is predicted that by 2025, the percentage science, molecular biology, genetics, and statistics,
4
of individuals living with CF will increase by 75%. The bioinformatics provides a tool to solve issues involving
incidence rate of CF in Africa and Asia is thought to be very molecular data by creating theoretical and computational
low. Nevertheless, the true prevalence report of this disease is models and tools. The escalating amount of biological and
5
often underestimated due to a lack of awareness, inadequate genetic data is managed using information technology
healthcare benefits, and a high infant mortality rate. 1 approaches, which entail collection, storing, analysis, and
integration of data. At present, bioinformatics is applied to a
Franconi and Anderson were the first to characterize wide range of significant tasks, such as analysis and prediction
CF, an autosomal recessive disorder, in 1936 and 1938, of the regulatory network of genes, gene expression, protein,
respectively. Knowledge about the CF transmembrane and gene structure, as well as functions and metabolic
6,7
conductance regulator (CFTR) gene, which is responsible pathways, to understand specific gene-disease relation.
8
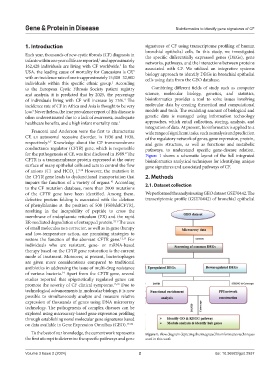
for the pathogenesis of CF, was first disclosed in 1989. The Figure 1 shows a schematic layout of the full integrated
CFTR is a transmembrane protein expressed at the outer bioinformatics analytical techniques for identifying unique
surface of many epithelial cells and acts to control the flow gene signatures and associated pathways of CF.
of anions (Cl and HCO ). However, the mutation in
−
− 9,10
3
the CFTR gene leads to dysfunctional transportation that 2. Methods
impairs the function of a variety of organs. According 2.1. Dataset collection
10
to the CF mutation database, more than 2000 mutations
of the CFTR gene have been identified. Among them, We performed the analysis using GEO dataset GSE70442. The
defective protein folding is associated with the deletion transcriptomic profile (GSE70442) of bronchial epithelial
of phenylalanine at the position of 508 (F508delCFTR),
resulting in the incapability of peptide to cross the
membrane of endoplasmic reticulum (ER) and the rapid
ER-mediated degradation of entrapped protein. The uses
7,11
of small molecules as a corrector, as well as in gene therapy
and low-temperature action, are promising strategies to
restore the function of the aberrant CFTR gene. 12,13 For
individuals who are resistant, gene- or mRNA-based
therapy based on the CFTR gene restoration is the current
mode of treatment. Moreover, at present, bacteriophages
are given more considerations compared to traditional
antibiotics in addressing the issue of multi-drug resistance
of various bacteria. Apart from the CFTR gene, several
14
studies reported that epigenetically regulated genes can
promote the severity of CF clinical symptoms. 15,16 Due to
technological advancements in molecular biology, it is now
possible to simultaneously analyze and measure relative
expression of thousands of genes using DNA microarray
technology. The pathogenesis of complex diseases can be
explored using microarray-based gene expression profiling
through establishing novel molecular gene signatures based
on data available in Gene Expression Omnibus (GEO). 17-22
To the best of our knowledge, the current work represents Figure 1. Flow diagram depicting the integrated bioinformatics techniques
the first attempt to determine the specific pathways and gene used in this work
Volume 3 Issue 2 (2024) 2 doi: 10.36922/gpd.2937