Page 133 - GPD-4-2
P. 133
Gene & Protein in Disease Binding of 11q to DENV and WNV proteases
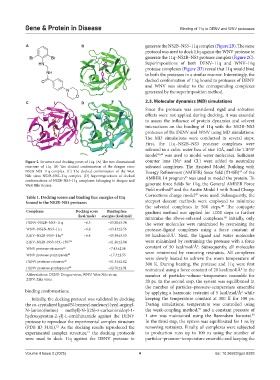
A B generate the NS2B–NS3–11q complex (Figure 2B). The same
protocol was used to dock 11q against the WNV protease to
generate the 11q–NS2B–NS3 protease complex (Figure 2C).
Superimpositions of both DENV–11q and WNV–11q
protease complexes (Figure 2D) reveal that 11q would bind
to both the proteases in a similar manner. Interestingly, the
docked conformation of 11q bound to proteases of DENV
and WNV was similar to the corresponding complexes
generated by the superimposition method.
C D 2.3. Molecular dynamics (MD) simulations
Since the protease was considered rigid and solvation
effects were not applied during docking, it was essential
to assess the influence of protein dynamics and solvent
interactions on the binding of 11q with the NS2B–NS3
proteases of the DENV and WNV using MD simulations.
The MD simulations were conducted in several steps.
First, the 11q–NS2B–NS3 protease complexes were
solvated in a cubic water box of size 10Å, and the TIP3P
model 43,44 was used to model water molecules. Sufficient
-
+
Figure 2. Structure and docking poses of 11q. (A) The two-dimensional counter ions (Na and Cl ) were added to neutralize
structure of 11q. (B) The docked conformation of the dengue virus solvated complexes. The Assisted Model Building with
NS2B–NS3–11q complex. (C) The docked conformation of the West Energy Refinement (AMBER) force field (ff14SB) of the
45
Nile virus NS2B–NS3–11q complex. (D) Superimpositions of docked AMBER 14 program was used to model the protein. To
45
conformations of NS2B–NS3–11q complexes belonging to dengue and
West Nile viruses. generate force fields for 11q, the General AMBER Force
Field method and the Austin Model 1 with Bond Charge
46
47
Table 1. Docking scores and binding free energies of 11q Corrections charge model were used. Subsequently, the
bound to the NS2B–NS3 proteases steepest descent methods were employed to minimize
the solvated complexes in 500 steps. The conjugate
48
Complexes Docking score Binding free gradient method was applied for 1,000 steps to further
(kcal/mole) energies (kcal/mol) minimize the above-solvated complexes. Initially, only
49
DENV–NS2B–NS3–11q −6.5 −15.80±3.34 the water molecules were minimized by restraining the
WNV–NS2B–NS3–11q −6.2 −13.13±2.56 protease–ligand complexes using a force constant of
2
ZIKV–NS2B–NS3–11q 34 −6.4 −15.59±3.53 50 kcal/mol/Å . Next, the ligand and water molecules
ZIKV–NS2B–NS3-SYC-1307 34 −11.26±2.84 were minimized by restraining the protease with a force
2
WNV protease-ritonavir 22 −7.43±2.16 constant of 50 kcal/mol/Å . Subsequently, all molecules
WNV protease-paritaprevir 22 −17.3±2.55 were minimized by removing restraints. All complexes
were slowly heated to achieve the room temperature of
DENV protease-ritonavir 22 −11.51±2.82
300 K. During heating, the protease and 11q were first
DENV protease-paritaprevir 22 −12.76±2.91 restrained using a force constant of 20 kcal/mol/Å in the
2
Abbreviations: DENV: Dengue virus; WNV: West Nile virus; number of particles–volume–temperature ensemble for
ZIKV: Zika virus. 20 ps. In the second step, the system was equilibrated in
the number of particles–pressure–temperature ensemble
binding conformations. by applying a harmonic restraint of 5 kcal/mol/Å while
2
Initially, the docking protocol was validated by docking keeping the temperature constant at 300 K for 100 ps.
the co-crystalized ligand N-benzoyl-norleucyl-lysyl-arginyl- During simulations, temperature was controlled using
50
N-[amino(imino) methyl]-N-[(2S)-5-carbamimidoyl-1- the weak-coupling method, and a constant pressure of
50
hydroxypentan-2-yl]-L-ornithinamide against the DENV 1 atm was maintained using the Barendsen barostat.
protease to reproduce the experimental complex structure In the third step, the system was equilibrated for 1 ns by
(PDB ID 3U1I). As the docking results reproduced the removing restraints. Finally, all complexes were subjected
12
experimental complex structure, the docking protocols to production runs up to 100 ns using the number of
12
were used to dock 11q against the DENV protease to particles–pressure–temperature ensemble and keeping the
Volume 4 Issue 2 (2025) 4 doi: 10.36922/gpd.8293