Page 135 - GPD-4-2
P. 135
Gene & Protein in Disease Binding of 11q to DENV and WNV proteases
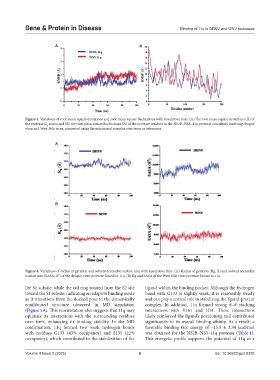
A B
Figure 3. Variations of root mean square deviations and root mean square fluctuations with simulation time. (A) The root mean square deviations (Å) of
the protease C atoms and (B) the root mean square fluctuations (Å) of the protease residues in the NS2B–NS3–11q protease complexes involving dengue
α
virus and West Nile virus, computed using the minimized complex structures as references.
A
B
Figure 4. Variations of radius of gyration and solvent accessible surface area with simulation time. (A) Radius of gyration (Rg, Å) and solvent accessible
2
surface area (SASA, Å ) of the dengue virus protease bound to 11q. (B) Rg and SASA of the West Nile virus protease bound to 11q.
the S1 subsite, while the tail ring rotated from the S2 site ligand within the binding pocket. Although the hydrogen
toward the S1 subsite, indicating an adaptive binding mode bond with G133 is slightly weak, it is reasonably steady
as it transitions from the docked pose to the dynamically and can play a critical role in stabilizing the ligand-protein
equilibrated structure observed in MD simulation complex. In addition, 11q formed strong π–π stacking
(Figure 5A). This reorientation also suggests that 11q may interactions with Y161 and H51. These interactions
optimize its interactions with the surrounding residues likely reinforced the ligand’s positioning and contributed
over time, enhancing its binding stability. In the MD significantly to its overall binding affinity. As a result, a
confirmation, 11q formed two weak hydrogen bonds favorable binding free energy of −15.8 ± 3.34 kcal/mol
with residues G133 (43% occupancy) and S135 (22% was obtained for the NS2B–NS3–11q protease (Table 1).
occupancy), which contributed to the stabilization of the This energetic profile supports the potential of 11q as a
Volume 4 Issue 2 (2025) 6 doi: 10.36922/gpd.8293