Page 97 - GTM-2-4
P. 97
Global Translational Medicine ECM receptor pathway in endotheliocytes after MI
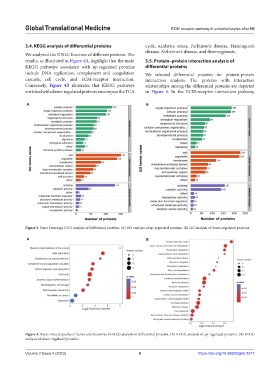
3.4. KEGG analysis of differential proteins cycle, oxidative stress, Parkinson’s disease, Huntington’s
disease, Alzheimer’s disease, and thermogenesis.
We analyzed the KEGG function of different proteins. The
results, as illustrated in Figure 4A, highlight that the main 3.5. Protein–protein interaction analysis of
KEGG pathways associated with up-regulated proteins differential proteins
include DNA replication, complement and coagulation We selected differential proteins for protein-protein
cascade, cell cycle, and ECM-receptor interaction. interaction analysis. The proteins with interaction
Conversely, Figure 4B illustrates that KEGG pathways relationships among the differential proteins are depicted
enriched with down-regulated proteins encompass the TCA in Figure 5. In the ECM-receptor interaction pathway,
A B
Figure 3. Gene Ontology (GO) analysis of differential proteins. (A) GO analysis of up-regulated proteins. (B) GO analysis of down-regulated proteins.
A B
Figure 4. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differential proteins. (A) KEGG analysis of up-regulated proteins. (B) KEGG
analysis of down-regulated proteins.
Volume 2 Issue 4 (2023) 6 https://doi.org/10.36922/gtm.2217