Page 98 - GTM-2-4
P. 98
Global Translational Medicine ECM receptor pathway in endotheliocytes after MI
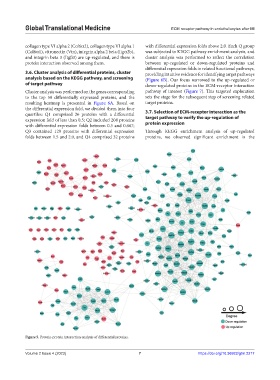
collagen type VI alpha 2 (Col6α2), collagen type VI alpha 1 with differential expression folds above 2.0. Each Q group
(Col6α1), vitronectin (Vtn), integrin alpha 2 beta (Itgα2b), was subjected to KEGG pathway enrichment analysis, and
and integrin beta 3 (Itgβ3) are up-regulated, and there is cluster analysis was performed to reflect the correlation
protein interaction observed among them. between up-regulated or down-regulated proteins and
differential expression folds in related functional pathways,
3.6. Cluster analysis of differential proteins, cluster providing intuitive evidence for identifying target pathways
analysis based on the KEGG pathway, and screening (Figure 6B). Our focus narrowed to the up-regulated or
of target pathway down-regulated proteins in the ECM-receptor interaction
Cluster analysis was performed on the genes corresponding pathway of interest (Figure 7). This targeted exploration
to the top 50 differentially expressed proteins, and the sets the stage for the subsequent step of screening related
resulting heatmap is presented in Figure 6A. Based on target proteins.
the differential expression fold, we divided them into four
quartiles: Q1 comprised 26 proteins with a differential 3.7. Selection of ECM-receptor interaction as the
expression fold of less than 0.5; Q2 included 208 proteins target pathway to verify the up-regulation of
with differential expression folds between 0.5 and 0.667; protein expression
Q3 contained 129 proteins with differential expression Through KEGG enrichment analysis of up-regulated
folds between 1.5 and 2.0, and Q4 comprised 32 proteins proteins, we observed significant enrichment in the
Figure 5. Protein-protein interaction analysis of differential proteins.
Volume 2 Issue 4 (2023) 7 https://doi.org/10.36922/gtm.2217