Page 294 - IJB-10-2
P. 294
International Journal of Bioprinting Kidney hydrogel print for renal cancer model
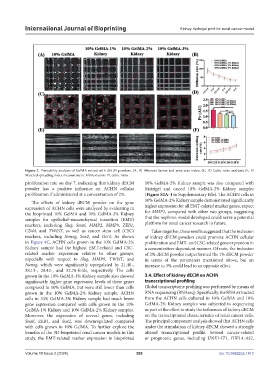
Figure 2. Printability analysis of GelMA mixed with dECM powders. (A, B) Filament fusion and pore area index; (C, D) Cubic ratio analysis; (E, F)
Material spreading index measurement. Abbreviation: Pr, cubic ratio.
proliferation rate on day 7, indicating that kidney dECM 10% GelMA-2% Kidney sample was also compared with
powder has a positive influence on ACHN cellular Matrigel and casted 10% GelMA-2% Kidney samples
proliferation if administered at a concentration of 2%. (Figure S2A–J in Supplementary File). The ACHN cells in
The effects of kidney dECM powder on the gene 10% GelMA-2% Kidney sample demonstrated significantly
expression of ACHN cells were analyzed by evaluating in higher expression for all EMT-related marker genes, expect
the bioprinted 10% GelMA and 10% GelMA-2% Kidney for MMP2, compared with other two groups, suggesting
samples for epithelial–mesenchymal transition (EMT) that the nephron model developed could serve a potential
markers, including Slug, Snail, MMP2, MMP9, ZEB1, platform for renal cancer research in future.
CD44, and TWIST, as well as cancer stem cell (CSC) Taken together, these results suggested that the inclusion
markers, including Nanog, Sox2, and Oct4. As shown of kidney dECM powders could promote ACHN cellular
in Figure 4C, ACHN cells grown in the 10% GelMA-2% proliferation and EMT- and CSC-related gene expression in
Kidney sample had the highest EMT-related and CSC- a concentration-dependent manner. Of note, the inclusion
related marker expression relative to other groups, of 2% dECM powder outperformed the 1% dECM powder
especially with respect to Slug, MMP9, TWIST, and in terms of the parameters mentioned above, but an
Nanog, which were significantly upregulated by 21.40-, increase to 3% could lead to an opposite effect.
38.13-, 20.42-, and 32.78-folds, respectively. The cells
grown in the 10% GelMA-1% Kidney sample also showed 3.4. Effect of kidney dECM on ACHN
significantly higher gene expression levels of these genes transcriptional profiling
compared to 10% GelMA, but were still lower than cells Global transcriptome profiling was performed by means of
grown in the 10% GelMA-2% Kidney sample. ACHN RNA sequencing (RNAseq). Specifically, the RNA extracted
cells in 10% GelMA-3% Kidney sample had much lower from the ACHN cells cultured in 10% GelMA and 10%
gene expression compared with cells grown in the 10% GelMA-2% Kidney samples was subjected to sequencing
GelMA-1% Kidney and 10% GelMA-2% Kidney samples. as part of the effort to study the influences of kidney dECM
Moreover, the expression of several genes, including on the transcriptional characteristics of renal cancer cells.
Snail, ZEB1, and Sox2, was downregulated compared The principal component analysis showed that ACHN cells
with cells grown in 10% GelMA. To further explore the under the stimulation of kidney dECM showed a strongly
benefits of the 3D-bioprinted renal cancer models in this altered transcriptional profile. Several cancer-related
study, the EMT-related marker expression in bioprinted or prognostic genes, including UNX1-IT1, HIF1A-AS2,
Volume 10 Issue 2 (2024) 286 doi: 10.36922/ijb.1413