Page 33 - IMO-2-3
P. 33
Innovative Medicines & Omics Tyrosine kinases: Structure, mechanism, and therapeutics
A B
C D
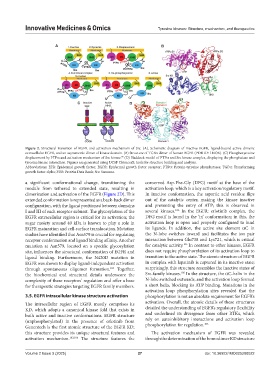
Figure 2. Structural transition of EGFR and activation mechanism of Src. (A). Schematic diagram of inactive EGFR, ligand-bound active dimeric
extracellular EGFR, and an asymmetric dimer of kinase domain. (B) Structure of TGFa dimer of human EGFR (PDB ID: 1MOX). (C) Phosphotyrosine
99
displacement by PTPa and activation mechanism of Src kinase. (D) Haddock model of PTPa and Src kinase complex, displaying the phosphatase and
tyrosine kinase interaction. Figures are generated using UCSF ChimeraX: tools for structure building and analysis.
Abbreviations: EFR: Epidermal growth factor; EGFR: Epidermal growth factor receptor; PTPa: Protein-tyrosine phosphatases; TGFa: Transforming
growth factor alpha; PDB: Protein Data Bank; Src: Sarcoma.
a significant conformational change, transitioning the conserved Asp-Phe-Gly (DFG) motif at the base of the
module from tethered to extended state, resulting in activation loop, which is a key activation/regulatory motif.
dimerization and activation of the EGFR (Figure 2D). This In inactive conformation, the aspartic acid residue flips
extended conformation is represented as a back-back dimer out of the catalytic center, making the kinase inactive
configuration, with the ligand positioned between domains and preventing the entry of ATP; this is observed in
106
I and III of each receptor subunit. The glycosylation of the several kinases. In the EGFR: erlotinib complex, the
EGFR extracellular region is critical for its activation; the DFG motif is found in the ‘in’ conformation; in this, the
sugar moiety around 40 kDa is known to play a role in activation loop is open and properly configured to bind
EGFR maturation and cell-surface translocation. Mutation its ligands. In addition, the active site element aC in
studies have identified that Asn579 is crucial for regulating the N-lobe switches inward and facilitates the ion pair
receptor conformation and ligand binding affinity. Another interaction between Glu738 and Lys721, which is critical
105
mutation at Asn579, located on a specific glycosylation for catalytic activity. In contrast to other kinases, EGFR
site, influences the structural conformation of EGFR and does not require phosphorylation of its activation loop to
ligand binding. Furthermore, the N420D mutation in transition to the active state. The atomic structure of EGFR
EGFR was shown to display ligand-independent activation in complex with lapatinib is captured in its inactive state;
through spontaneous oligomer formation. Together, surprisingly, this structure resembles the inactive states of
105
105
the biochemical and structural details underscore the Src-family kinases. In the structure, the aC-helix in the
complexity of these receptors’ regulation and offer a base N-lobe switched outwards, and the activation loop formed
for therapeutic strategies targeting EGFR family members. a short helix, blocking its ATP binding. Mutations in the
activation loop phosphorylation sites revealed that the
3.5. EGFR intracellular kinase structure activation phosphorylation is not an absolute requirement for EGFR’s
The intracellular region of EGFR mostly comprises its activation. Overall, the atomic details of these structures
KD, which adopts a canonical kinase fold that exists in detailed the understanding of EGFR’s regulatory flexibility
both active and inactive conformations. EGFR structure and underlined its divergence from other RTKs, which
(unphosphorylated) in the presence of erlotinib from rely on autoinhibitory interactions and activation loop
105
Genentech is the first atomic structure of the EGFR KD; phosphorylation for regulation.
this structure provides its unique structural features and The activation mechanism of EGFR was revealed
activation mechanism. 103,106 The structure features the through the determination of the homodimer KD structure
Volume 2 Issue 3 (2025) 27 doi: 10.36922/IMO025200022