Page 82 - ITPS-7-1
P. 82
INNOSC Theranostics and
Pharmacological Sciences PfHSP and polyamines interactions
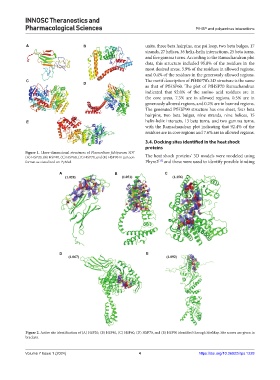
A B units, three beta hairpins, one psi loop, two beta bulges, 17
strands, 27 helices, 36 helix-helix interactions, 25 beta turns,
and five gamma turns. According to the Ramachandran plot
data, this structure included 95.8% of the residues in the
most desired areas, 3.9% of the residues in allowed regions,
and 0.4% of the residues in the generously allowed regions.
C The motif description of PfHSP70’s 3D structure is the same
D
as that of PfHSP60. The plot of PfHSP70 Ramachandran
indicated that 92.0% of the amino acid residues are in
the core areas, 7.3% are in allowed regions, 0.5% are in
generously allowed regions, and 0.2% are in banned regions.
The generated PfHSP90 structure has one sheet, four beta
hairpins, two beta bulges, nine strands, nine helices, 15
E helix-helix interacts, 13 beta turns, and two gamma turns,
with the Ramachandran plot indicating that 92.4% of the
residues are in core regions and 7.6% are in allowed regions.
3.4. Docking sites identified in the heat shock
proteins
Figure 1. Three-dimensional structures of Plasmodium falciparum 3D7
(A) HSP20, (B) HSP40, (C) HSP60, (D) HSP70, and (E) HSP90 in cartoon The heat shock proteins’ 3D models were modeled using
[15]
format as visualized on PyMol. Phyre2 and these were used to identify possible binding
A B C
D E
Figure 2. Active site identification of (A) HSP20, (B) HSP40, (C) HSP60, (D) HSP70, and (E) HSP90 identified through SiteMap. Site scores are given in
brackets.
Volume 7 Issue 1 (2024) 4 https://doi.org/10.36922/itps.1228