Page 83 - ITPS-7-1
P. 83
INNOSC Theranostics and
Pharmacological Sciences PfHSP and polyamines interactions
A B C
D E
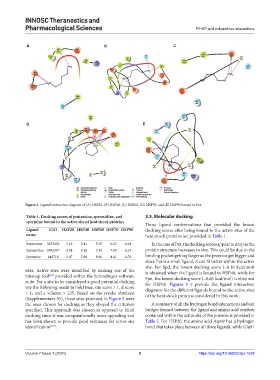
Figure 3. Ligand interaction diagram of (A) HSP20, (B) HSP40, (C) HSP60, (D) HSP70, and (E) HSP90 bound to Put.
Table 1. Docking scores of putrescine, spermidine, and 3.5. Molecular docking
spermine bound to the active site of heat shock proteins
Those ligand conformations that provided the lowest
Ligand CID HSP20 HSP40 HSP60 HSP70 HSP90 docking scores after being bound to the active sites of the
name heat shock proteins are provided in Table 1.
Putrescine 3452892 −3.19 −5.41 −5.95 −6.63 −6.68 In the case of Put, the docking scores appear to drop as the
Spermidine 6992097 −4.54 −8.18 −7.34 −7.99 −6.19 protein structure increases in size. This could be due to the
Spermine 446718 −3.07 −5.86 −8.66 −4.41 −4.76 binding pocket getting larger as the proteins get bigger and
since Put is a small ligand, it can fit better within the active
site. For Spd, the lowest docking score (–8.18 kcal/mol)
sites. Active sites were identified by making use of the is obtained when the ligand is bound to HSP40, while for
Sitemap tool provided within the Schrodinger software Spn, the lowest docking score (–8.66 kcal/mol) is obtained
[18]
suite. For a site to be considered a good potential docking for HSP60. Figures 3–5 provide the ligand interaction
site the following needs to hold true, site score > 1, d score diagrams for the different ligands bound to the active sites
> 1, and a volume > 225. Based on the results obtained of the heat shock proteins considered in this work.
(Supplementary S5), those sites provided in Figure 2 were
the ones chosen for docking as they obeyed the criterion A summary of all the hydrogen bond interactions and salt
specified. This approach was chosen as opposed to blind bridges formed between the ligand and amino acid residues
docking since it was computationally more appealing and contained within the active site of the proteins is provided in
has been shown to provide good estimates for active site Table 2. For HSP20, the amino acid Asp89 has a hydrogen
identification [6,7] . bond that takes place between all three ligands, while Glu91
Volume 7 Issue 1 (2024) 5 https://doi.org/10.36922/itps.1228