Page 86 - ITPS-7-1
P. 86
INNOSC Theranostics and
Pharmacological Sciences PfHSP and polyamines interactions
A B
C D
E
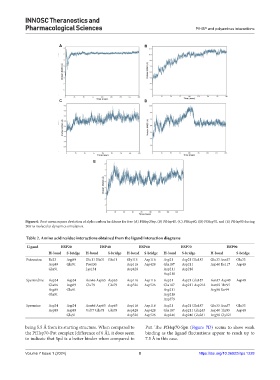
Figure 6. Root mean square deviation of alpha carbon backbone for free (A) PfHsp20sp, (B) PfHsp40, (C) PfHsp60, (D) PfHsp70, and (E) PfHsp90 during
200 ns molecular dynamics simulation.
Table 2. Amino acid residue interactions obtained from the ligand interaction diagrams
Ligand HSP20 HSP40 HSP60 HSP70 HSP90
H-bond S-bridge H-bond S-bridge H-bond S-bridge H-bond S-bridge H-bond S-bridge
Putrescine Ile23 Asp89 Glu11 His31 Glu11 Gly115 Asp116 Asp21 Asp21 Glu187 Glu33 Asn37 Glu33
Asp89 Glu91 Pro130 Asp116 Asp428 Glu187 Asp211 Asp40 Ile117 Asp40
Glu91 Lys134 Asp428 Asp211 Asp218
Asp218
Spermidine Asp24 Asp24 Asn46 Asp65 Asp65 Asp116 Asp116 Asp21 Asp21 Glu187 Asn37 Asp40 Asp40
Glu86 Asp89 Glu79 Glu79 Asp526 Asp526 Glu187 Asp211 Asp218 Asn92 Thr95
Asp89 Glu91 Asp211 Arg98 Ser99
Glu91 Asp218
Asp379
Spermine Asp24 Asp24 Asn46 Asp65 Asp65 Asp116 Asp116 Asp21 Asp21 Glu187 Glu33 Asn37 Glu33
Asp89 Asp89 Val77 Glu78 Glu79 Asp428 Asp428 Glu187 Asp211 Glu243 Asp40 Thr95 Asp40
Glu91 Asp526 Asp526 Asp246 Asp246 Glu281 Arg98 Gly121
being 5.5 Å from its starting structure. When compared to Put. The PfHsp70-Spn (Figure 7D) seems to show weak
the PfHsp70-Put complex (difference of 6 Å), it does seem binding as the ligand fluctuations appear to reach up to
to indicate that Spd is a better binder when compared to 7.5 Å in this case.
Volume 7 Issue 1 (2024) 8 https://doi.org/10.36922/itps.1228