Page 84 - ITPS-7-1
P. 84
INNOSC Theranostics and
Pharmacological Sciences PfHSP and polyamines interactions
has a hydrogen bond present for ligands Put and Spd. Asp24 produced during the MD simulation. Figure 6 does show
shows a hydrogen bond between Spd and Spn. It is this that PfHsp20, PfHsp40, and PfHsp70 stabilize within the
combination of salt bridges and hydrogen bond interactions 200 ns, but PfHsp60 and PfHsp90 appeared to have some
that adds to the protein-ligand complex stability. excess fluctuations towards the end of the 200 ns. Although
this was the case, it was decided that we would make use
3.6. MD of a maximum of 200 ns for the protein-ligand complex
Before we investigated the results obtained for the various HSP simulations due to the limited resources that we had access
complexes, we first needed to make sure that the environment to at the high-performance computing center.
chosen for our MD studies was appropriate. Figure 6 shows MD simulations of the Hsps with the polyamines
the root mean square deviation (RMSD) results obtained for showed that putrescine best binds to PfHsp60 and PfHsp70
200 ns simulations of (A) HSP20, (B) HSP40, (C) HSP60, (D) (Figure 7) compared to the other Hsps. Put seems to remain
HSP70, and (E) HSP90 when there were no ligands present strongly bound to the active site of PfHsp60 for around 125
within the active site of the proteins. ns before the ligand begins to drift away from the active
The RMSD is used for measuring the changes that take place site, reaching distances of up to 14 Å from the starting
in the protein while present in a solvated environment . structure (Figure 7A). For ligands to be considered good
[25]
A protein’s stability can be determined by the deviations inhibitors, they need to remain bound to the active site for
A B C
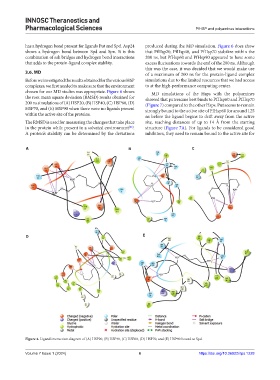
D E
Figure 4. Ligand interaction diagram of (A) HSP20, (B) HSP40, (C) HSP60, (D) HSP70, and (E) HSP90 bound to Spd.
Volume 7 Issue 1 (2024) 6 https://doi.org/10.36922/itps.1228