Page 80 - ITPS-8-3
P. 80
INNOSC Theranostics and
Pharmacological Sciences Activity of green-synthesized nanoparticles
Figure 1. Ultraviolet-visible spectra of biosynthesized silver nanoparticles from carpenter bee wing extract
silver nanoparticles (Figure 2). In addition, SEM A B
analysis shows that the nanoparticles exhibit a spherical
morphology with an approximate diameter ranging from
10.0 nm to 40.0 nm (Figure 2).
A decrease in optical density with increasing nanoparticle
concentration suggests that silver nanoparticles inhibit the
growth of E. coli and K. pneumoniae (Figure 3). To further
investigate the bacterial response to silver nanoparticles,
stress indicators, cell morphology, and nanoparticle–cell
interactions were examined using SEM. The results show C D
no aggregation in the control samples (i.e., in the absence
of nanoparticles) (Figures 4A-D and 5A-D). However,
aggregation is observed in E. coli and K. pneumoniae cells
treated with silver nanoparticles (Figures 4E-H and 5E-H).
3.2. Genomic analysis
WGS analysis was conducted on control and treated
K. pneumoniae and E. coli cells and compared against
their respective reference genomes to identify potential
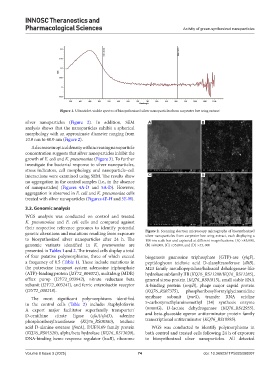
genetic alterations and mutations resulting from exposure Figure 2. Scanning electron microscopy micrographs of biosynthesized
silver nanoparticles from carpenter bee wing extract, each displaying a
to biosynthesized silver nanoparticles after 24 h. The 100 nm scale bar and captured at different magnifications: (A) ×43,000,
genomic variants identified in K. pneumoniae are (B) ×60,000, (C) ×35,000, and (D) ×43, 000
presented in Tables 1 and 2. The treated cells display a total
of four putative polymorphisms, three of which exceed biogenesis guanosine triphosphate (GTP)-ase (ylqF),
a frequency of 0.5 (Table 1). These include mutations in peptidoglycan teichoic acid D-alanyltransferase (dltB),
the putrescine transport system adenosine triphosphate M23 family metallopeptidase/haloacid dehalogenase-like
(ATP)-binding protein (J2Y72_004072), multidrug (MDR) hydrolase subfamily IIB (KQ76_RS11280/KQ76_RS11285),
efflux pump (J2Y72_003942), nitrate reductase beta general stress protein (KQ76_RS01815), small stable RNA
subunit (J2Y72_003241), and ferric enterobactin receptor A-binding protein (smpB), phage major capsid protein
(J2Y72_000218). (KQ76_RS07375), phosphoribosylformylglycinamidine
The most significant polymorphisms identified synthase subunit (purS), transfer RNA uridine
in the control cells (Table 2) include: Staphyloferrin 5-carboxymethylaminomethyl (34) synthesis enzyme
A export major facilitator superfamily transporter/ (mnmG), D-lactate dehydrogenase (KQ76_RS12955),
D-ornithine citrate ligase (sfaA/sfaD), adenine and beta-glucoside operon antiterminator protein family
phosphoribosyltransferase (KQ76_RS08360), teichoic transcriptional antiterminator (KQ76_RS10985).
acid D-alanine esterase (fmtA), DUF3169 family protein WGS was conducted to identify polymorphisms in
(KQ76_RS01520), alpha/beta hydrolase (KQ76_RS13020), both control and treated cells following 24 h of exposure
DNA-binding heme response regulator (hssR), ribosome to biosynthesized silver nanoparticles. All detected
Volume 8 Issue 3 (2025) 74 doi: 10.36922/ITPS025080007