Page 123 - MI-2-4
P. 123
Microbes & Immunity Isolation and identification of PEDV strain CHN-CQ-2021
was observed in Vero cells at 8 hpi. Prolonged incubation gene sequences of other reference strains retrieved from
resulted in progressive expansion of the cytopathic area GenBank. Sequence alignment and phylogenetic analysis
and the emergence of cell detachment (Figure 3A). were performed using the neighbor-joining method
Analysis of the virus proliferation curve further revealed to construct a phylogenetic tree, with bootstrap values
that CHN-CQ-2021 reached its replication peak in Vero applied to evaluate the reliability of the tree topology. The
28
cells at 16 hpi (Figure 3B). These results demonstrate that PEDV CHN-CQ-2021 strain was classified within the G2b
the PEDV CHN-CQ-2021 strain can efficiently proliferate subgroup and displayed the closest genetic relationship to
in vitro. strain GD-1 (GenBank: JX647847). In contrast, it exhibited
significant phylogenetic divergence from the G1-type
3.3. Phylogenetic analysis of whole-genome and S
genes of PEDV CHN-CQ-2021 classical strain branch represented by CV777 (Figure 4).
These data suggested that the PEDV CHN-CQ-2021 strain
To investigate the genetic evolution of the strain, the was the most closely related to other PEDV strains from
complete genome of PEDV CHN-CQ-2021 was amplified mainland China.
using RT-PCR and compared with the full genome and S
3.4. High pathogenicity of PEDV CHN-CQ-2021
strain in newborn piglets
To evaluate the pathogenicity of the PEDV CHN-CQ-2021
strain in piglets, newborn piglets were orally infected
with the CHN-CQ-2021 strain. Compared to the control
group, CHN-CQ-2021 strain-infected piglets exhibited
typical clinical symptoms, including severe watery
diarrhea and dehydration (Figure 5). Importantly, all
CHN-CQ-2021 strain-infected newborn piglets died
within 4 days post-inoculation (Figure 6). Further
analysis of virus shedding and tissue tropism indicated
that, compared to the control group, CHN-CQ-2021
strain-infected newborn piglets shed virus at varying
levels during the 4-day observation period (Figure 7A),
while viral nucleic acids were detected in both intestinal
and brain tissues (Figure 7B). These results suggest that
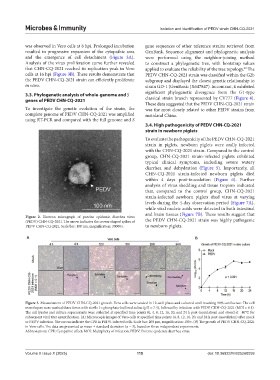
Figure 2. Electron micrograph of porcine epidemic diarrhea virus
(PEDV) CHN-CQ-2021. The arrow indicates the crown-shaped spikes of the PEDV CHN-CQ-2021 strain was highly pathogenic
PEDV CHN-CQ-2021. Scale bar: 100 nm, magnification: 30000×. to newborn piglets.
A B
Figure 3. Measurement of PEDV CHN-CQ-2021 growth. Vero cells were seeded in 12-well plates and cultured until reaching 90% confluence. The cell
monolayers were washed three times with sterile 1× phosphate-buffered saline (pH = 7.4), followed by infection with PEDV CHN-CQ-2021 (MOI = 0.1).
The cell lysates and culture supernatants were collected at specified time points (0, 4, 8, 12, 16, 20, and 24 h post-inoculation) and stored at −80°C for
subsequent viral titer quantification. (A) Microscopic images of Vero cells at specified time points (4, 8, 12, 16, 20, and 24 h post-inoculation) after mock
or PEDV infection. The arrows indicate the CPE in PEDV-infected cells. Scale bar: 200 μm, magnification: 100×. (B) The growth of PEDV CHN-CQ-2021
in Vero cells. The data are presented as mean ± standard deviation (n = 3), based on three independent experiments.
Abbreviations: CPE: Cytopathic effect; MOI: Multiplicity of infection; PEDV: Porcine epidemic diarrhea virus.
Volume X Issue X (2025) 115 doi: 10.36922/MI025260059