Page 41 - TD-3-3
P. 41
Tumor Discovery LncRNA HA117 in osteosarcoma regulation
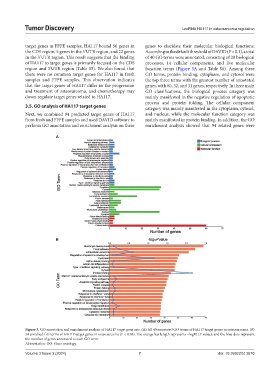
target genes in FFPE samples, HA117 bound 56 genes in genes to elucidate their molecular biological functions.
the CDS region, 5 genes in the 5’UTR region, and 22 genes According to the default threshold of DAVID (P < 0.1), a total
in the 3’UTR region. This result suggests that the binding of 40 GO terms were annotated, consisting of 20 biological
of HA117 to target genes is primarily focused on the CDS processes, 14 cellular components, and five molecular
region and 3’UTR region (Table S5). We also found that function terms (Figure 3A and Table S6). Among these
there were no common target genes for HA117 in fresh GO terms, protein binding, cytoplasm, and cytosol were
samples and FFPE samples. This observation indicates the top three terms with the greatest number of annotated
that the target genes of HA117 differ in the progression genes, with 60, 32, and 31 genes, respectively. In three main
and treatment of osteosarcoma, and chemotherapy may GO classifications, the biological process category was
down-regulate target genes related to HA117. mainly manifested in the negative regulation of apoptotic
process and protein folding. The cellular component
3.5. GO analysis of HA117 target genes category was mainly manifested in the cytoplasm, cytosol,
Next, we combined 94 predicted target genes of HA117 and nucleus, while the molecular function category was
from fresh and FFPE samples and used DAVID software to mainly manifested in protein binding. In addition, the GO
perform GO annotation and enrichment analysis on these enrichment analysis showed that 94 related genes were
A
B
Figure 3. GO annotation and enrichment analysis of HA117 target gene sets. (A) All 40 annotated GO terms of HA117 target genes in osteosarcoma. (B)
24 enriched GO terms of HA117 target genes in osteosarcoma (P < 0.05). The orange bar length represents −log10 (P-value), and the blue dots represent
the number of genes annotated to each GO term.
Abbreviation: GO: Gene ontology.
Volume 3 Issue 3 (2024) 7 doi: 10.36922/td.3670