Page 40 - TD-3-3
P. 40
Tumor Discovery LncRNA HA117 in osteosarcoma regulation
to the analysis results of fresh samples, in FFPE samples, coefficient rho >0.8 and P < 0.05. Only HA117 and other
HA117 also showed down-regulation with chemotherapy, mRNAs meeting these conditions were recognized as
but not significantly (P > 0.05, Figure 1D). In summary, having an interaction relationship. In fresh samples, we
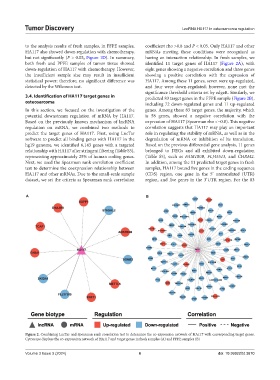
both fresh and PPFE samples of tumor tissue showed identified 11 target genes of HA117 (Figure 2A), with
down-regulation of HA117 with chemotherapy. However, eight genes showing a negative correlation and three genes
the insufficient sample size may result in insufficient showing a positive correlation with the expression of
statistical power; therefore, no significant difference was HA117. Among these 11 genes, seven were up-regulated,
detected by the Wilcoxon test. and four were down-regulated; however, none met the
significance threshold criteria set by edgeR. Similarly, we
3.4. Identification of HA117 target genes in predicted 83 target genes in the FFPE sample (Figure 2B),
osteosarcoma including 72 down-regulated genes and 11 up-regulated
In this section, we focused on the investigation of the genes. Among these 83 target genes, the majority, which
potential downstream regulation of mRNA by HA117. is 58 genes, showed a negative correlation with the
Based on the previously known mechanism of lncRNA expression of HA117 (Spearman rho <−0.8). This negative
regulation on mRNA, we combined two methods to correlation suggests that HA117 may play an important
predict the target genes of HA117. First, using LncTar role in regulating the stability of mRNA, as well as in the
software to predict all binding genes with HA117 in the degradation of mRNA or inhibition of its translation.
hg19 genome, we identified 6,143 genes with a targeted Based on the previous differential gene analysis, 11 genes
relationship with HA117 after stringent filtering (Table S5), belonged to DEGs and all exhibited down-regulation
representing approximately 25% of human coding genes. (Table S5), such as FAM180B, FLJ45513, and CHRM2.
Next, we used the Spearman rank correlation coefficient In addition, among the 11 predicted target genes in fresh
test to determine the coexpression relationship between samples, HA117 bound five genes in the coding sequence
HA117 and other mRNAs. Due to the small-scale sample (CDS) region, one gene in the 5’ untranslated (UTR)
dataset, we set the criteria as Spearman rank correlation region, and five genes in the 3’UTR region. For the 83
A B
Figure 2. Combining LncTar and Spearman rank correlation test to determine the co-expression network of HA117 with corresponding target genes.
Cytoscape displays the co-expression network of HA117 and target genes in fresh samples (A) and FFPE samples (B)
Volume 3 Issue 3 (2024) 6 doi: 10.36922/td.3670