Page 158 - TD-3-4
P. 158
Tumor Discovery CK2 deregulation in non-small-cell lung cancer
A
B C
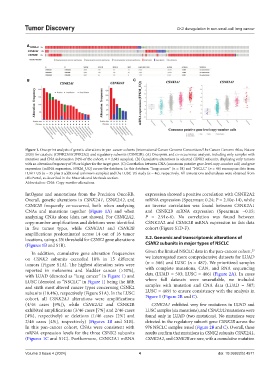
Figure 1. Oncoprint analysis of genetic alterations in pan-cancer cohorts (International Cancer Genome Consortium/The Cancer Genome Atlas, Nature
2020) for catalytic (CSNK2A1/CSNK2A2) and regulatory subunits (CSNK2B). (A) Oncoprint and co-occurrence analysis, including only samples with
mutation and CNA information (92% of the cohort, n = 2,683 samples). (B) Cumulative alterations in selected CSNK2 subunits, displaying only tumors
with an alteration frequency of 3% or higher for the target gene. (C) Correlation between CNA (consensus putative gene-level copy-number call) and gene
expression (mRNA expression, FPKM_UQ) across the database. In this database, “lung cancer” (n = 38) and “NSCLC” (n = 46) encompass data from
LUAD-US (n = 35 plus 3 additional unknown samples) and the LUSC-US study (n = 46), respectively. All annotations and analyses were obtained from
cBioPortal, as described in the Materials and Methods section.
Abbreviation: CNA: Copy-number alterations.
IntOgene and annotations from the Precision OncoKB. expression showed a positive correlation with CSNK2A2
Overall, genetic alterations in CSNK2A1, CSNK2A2, and mRNA expression (Spearman: 0.24; P = 2.10e-14), while
CSNK2B frequently co-occurred, both when analyzing an inverse correlation was found between CSNK2A1
CNAs and mutations together (Figure 1A) and when and CSNK2B mRNA expression (Spearman: −0.15;
analyzing CNAs alone (data not shown). For CSNK2A2, P = 2.91e-6). No correlation was found between
copy-number amplifications and deletions were identified CSNK2A2 and CSNK2B mRNA expression in this data
in five tumor types, while CSNK2A1 and CSNK2B cohort (Figure S1D-F).
amplifications predominated across 14 out of 16 tumor
locations, using a 3% threshold for CSNK2 gene alterations 3.2. Genomic and transcriptomic alterations of
(Figures 1B and S1B). CSNK2 subunits in major types of NSCLC
23
In addition, cumulative gene alteration frequencies Given the limited NSCLC data in the pan-cancer cohort,
on CSNK2 subunits exceeded 10% in 15 different we interrogated more comprehensive datasets for LUAD
tumors (Figure S1A). The highest alteration rates were (n = 566) and LUSC (n = 487). We prioritized samples
reported in melanoma and bladder cancer (>30%), with complete mutations, CAN, and RNA sequencing
with LUAD (denoted as “lung cancer” in Figure 1) and data (LUAD = 503; LUSC = 466) (Figure 2A). In cases
LUSC (denoted as “NSCLC” in Figure 1) being the fifth where full datasets were unavailable, we included
and sixth most altered cancer types concerning CSNK2 samples with mutation and CNA data (LUAD = 507;
subunits (18.4%), respectively (Figure S1A). In the LUSC LUSC = 469) to ensure consistency with the analysis in
cohort, all CSNK2A1 alterations were amplifications Figure 1 (Figure 2B and C).
(4/46 cases [9%]), while CSNK2A2 and CSNK2B CSNK2A1 exhibited very few mutations in LUAD and
exhibited amplifications (3/46 cases [7%] and 2/46 cases LUSC samples (six mutations), and CSNK2A2 mutations were
[4%], respectively) or deletions (1/46 cases [2%] and found only in LUAD (two mutations). No mutations were
2/46 cases [4%], respectively) (Figures 1B and S1B). detected in the regulatory subunit gene CSNK2B across the
In this pan-cancer cohort, CNAs were consistent with 976 NSCLC samples tested (Figure 2B and C). Overall, these
mRNA expression levels for the three CSNK2 subunits results confirm that mutations in CSNK2 subunits CSNK2A1,
(Figures 1C and S1C). Furthermore, CSNK2A1 mRNA CSNK2A2, and CSNK2B are rare, with a cumulative mutation
Volume 3 Issue 4 (2024) 4 doi: 10.36922/td.4571