Page 162 - TD-3-4
P. 162
Tumor Discovery CK2 deregulation in non-small-cell lung cancer
A B
C
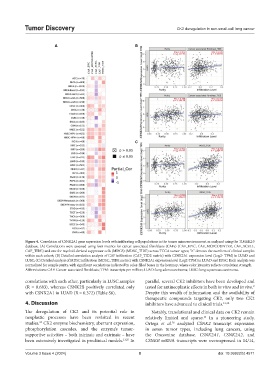
Figure 4. Correlation of CSNK2A1 gene expression levels with infiltrating cell populations in the tumor microenvironment, as analyzed using the TIMER2.0
database. (A) Correlations were assessed using four metrics for cancer-associated fibroblasts (CAFs) (CAF_EPIC, CAF_MCPCOUNTER, CAF_XCELL,
CAF_TIDE) and one for myeloid-derived suppressor cells (MDCS) (MDSC_TIDE) across TCGA tumor types; “n” denotes the numbers of clinical samples
within each cohort; (B) Detailed correlation analysis of CAF infiltration (CAF_TIDE metric) with CSNK2A1 expression level (Log2- TPM) in LUAD and
LUSC; (C) Detailed analysis of MDSC infiltration (MDSC_TIDE metric) with CSNK2A1 expression level (Log2-TPM) in LUAD and LUSC. Each analysis was
normalized for sample purity, with significant correlations indicated by color-filled boxes in the heatmap, where color intensity reflects correlation strength.
Abbreviations: CAF: Cancer-associated fibroblasts; TPM: transcripts per million; LUAD: lung adenocarcinoma; LUSC: lung squamous carcinoma.
correlations with each other, particularly in LUSC samples parallel, several CK2 inhibitors have been developed and
(R = 0.683), whereas CSNK2B positively correlated only tested for antineoplastic effects in both in vitro and in vivo.
9
with CSNK2A1 in LUAD (R = 0.372) (Table S6). Despite this wealth of information and the availability of
therapeutic compounds targeting CK2, only two CK2
4. Discussion inhibitors have advanced to clinical trials. 11,12
The deregulation of CK2 and its potential role in Notably, translational and clinical data on CK2 remain
neoplastic processes have been revisited in recent relatively limited and sparse. In a pioneering study,
13
studies. CK2 enzyme biochemistry, aberrant expression, Ortega et al. analyzed CSNK2 transcript expression
24
26
phosphorylation cascades, and the enzyme’s tumor- in seven tumor types, including lung cancers, using
supportive activities – both intrinsic and extrinsic – have the Oncomine database. CSNK2A1, CSNK2A2, and
been extensively investigated in preclinical models. 13,25 In CSNKB mRNA transcripts were overexpressed in 14/14,
Volume 3 Issue 4 (2024) 8 doi: 10.36922/td.4571