Page 32 - AN-1-2
P. 32
Advanced Neurology Role of Panax ginseng in neuronal complications
with the significance level set at 0.05. The parameters
set for this analysis were medium network and two-
sided hypergeometric test with Bonferroni correction.
Eventually, the functional network was analyzed and
visualized using algorithmic organic layout.
2.5. Docking study
Four compounds (gensinside, panaxydol, panaxynol, and
panaxytriol) were chosen to dock with the selected targets
(Glutathione S-transferase P1 [GSTP1], HSP90, and
Mitogen-activated protein kinases [MAPK8]) in search of
a suitable ligand that could match and favorably bind with
the protein . The preparation of ligands was performed
[27]
by searching the PubChem database, after which the ligand
structures were saved in mdl. format (v3000). This file was
then visualized in AutoDock (version 4.2). The non-polar H
2
atoms were combined while the torsion value was defined
to default. Kollman and Gasteiger charges were affixed
with the aid of default parameters. To deduce the function
of the acquired protein targets in neuronal damage, three-
dimensional structure of the acquired protein targets was
downloaded from the PDB database in.pdb format (www.rcsb.
org). The structure was visualized in AutoDock (version 4.2),
after which the H O molecules and the polar H atoms were
2
2
deleted and added, respectively. The same software was used
to dock the compounds with GSTP1, HSP90, and MAPK8.
All the compounds were individually docked with the same
ligand, after the grid-box for all compounds was assigned
before docking, where grid parameters were established
to affirm the binding of ligands to the binding sites of
protein, and the output was saved in the GPF format. The
number of runs was adjusted to 30, while Lamarckian-GA
(version 4.2) was chosen for processing the output file. The
binding energy was selected as the main mode of result
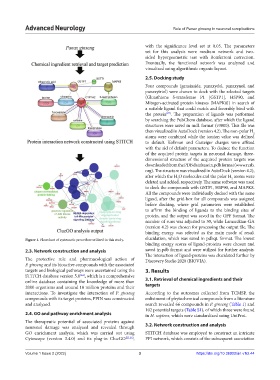
Figure 1. Flowchart of systematic procedure utilized in this study. elucidation, which was saved in pdbqt. format. The lowest
binding energy scores of ligand-proteins were chosen and
2.3. Network construction and analysis saved in.pdb format and were utilized for further analysis.
The interaction of ligand-proteins was elucidated further by
The protective role and pharmacological action of Discovery Studio 2020 (BIOVIA).
P. ginseng and its bioactive compounds with the associated
targets and biological pathways were ascertained using the 3. Results
STITCH database version 5.0 , which is a comprehensive
[24]
online database containing the knowledge of more than 3.1. Retrieval of chemical ingredients and their
2000 organisms and around 10 million proteins and their targets
interactions. To investigate the interaction of P. ginseng According to the outcomes collected from TCMSP, the
compounds with its target proteins, PPIN was constructed enlistment of phytochemical compounds from a literature
and analyzed. search revealed 66 compounds in P. ginseng (Table 1) and
102 potential targets (Table S1), of which three were found
2.4. GO and pathway enrichment analysis in H. sapiens, which were standardized using UniProt.
The therapeutic potential of associated proteins against
neuronal damage was analyzed and revealed through 3.2. Network construction and analysis
GO enrichment analysis, which was carried out using STITCH database was employed to construct an intricate
Cytoscape (version 3.4.0) and its plug-in ClueGO [25,26] , PPI network, which consists of the subsequent association
Volume 1 Issue 2 (2022) 3 https://doi.org/10.36922/an.v1i2.44