Page 100 - GPD-2-1
P. 100
Gene & Protein in Disease SCN7A is a protective factor in LUAD
A B
C
D
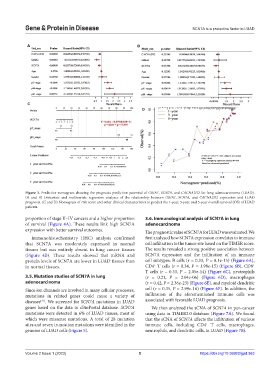
Figure 3. Predictive nomogram showing the prognosis prediction potential of GRIA1, SCN7A, and CACNA2D2 for lung adenocarcinoma (LUAD).
(A and B) Univariate and multivariate regression analyses of the relationship between GRIA1, SCN7A, and CACNA2D2 expression and LUAD
prognosis. (C and D) Nomogram of risk score and other clinical characteristics to predict the 1-year, 3-year, and 5-year overall survival (OS) of LUAD
patients.
proportion of stage II–IV cancers and a higher proportion 3.6. Immunological analysis of SCN7A in lung
of survival (Figure 4A). These results link high SCN7A adenocarcinoma
expression with better survival outcomes. The prognostic value of SCN7A for LUAD was evaluated. We
Immunohistochemistry (IHC) analysis confirmed first analyzed how SCN7A expression correlates to immune
that SCN7A was moderately expressed in normal cell infiltration to the tumor site based on the TIMER score.
tissues but was entirely absent in lung cancer tissues The results revealed a strong positive association between
(Figure 4B). These results showed that mRNA and SCN7A expression and the infiltration of six immune
protein levels of SCN7A are lower in LUAD tissues than cell subtypes: B cells (r = 0.33, P = 8.1e-15) (Figure 6A),
+
+
in normal tissues. CD4 T cells (r = 0.34, P = 1.98e-15) (Figure 6B), CD8
T cells (r = 0.33, P = 2.05e-14) (Figure 6C), neutrophils
3.5. Mutation studies of SCN7A in lung (r = 0.21, P = 2.04e-06) (Figure 6D), macrophages
adenocarcinoma (r = 0.42, P = 2.36e-23) (Figure 6E), and myeloid dendritic
Since ion channels are involved in many cellular processes, cell (r = 0.35, P = 2.99e-16) (Figure 6F). In addition, the
mutations in related genes could cause a variety of infiltration of the aforementioned immune cells was
[21]
diseases . We screened for SCN7A mutations in LUAD associated with favorable LUAD prognosis.
genes based on the data in cBioPortal database. SCN7A We then analyzed the sCNA of SCN7A in pan-cancer
mutations were detected in 6% of LUAD tissues, most of using data in TIMER2.0 database (Figure 7A). We found
which were missense mutations. A total of 28 mutation that the sCNA of SCN7A affects the infiltration of various
sites and seven truncation mutations were identified in the immune cells, including CD4 T cells, macrophages,
+
genome of LUAD cells (Figure 5). neutrophils, and dendritic cells, in LUAD (Figure 7B).
Volume 2 Issue 1 (2023) 5 https://doi.org/10.36922/gpd.363