Page 35 - GTM-1-2
P. 35
Global Translational Medicine Proteomic analysis of heart in metabolic cardiomyopathy
A B
C
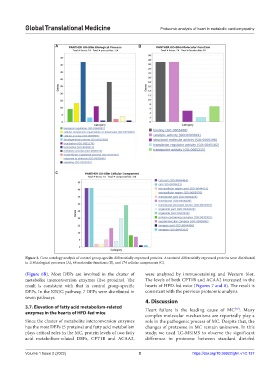
Figure 3. Gene ontology analysis of control group-specific differentially expressed proteins. Annotated differentially expressed proteins were distributed
in 114 biological processes (A), 68 molecular functions (B), and 174 cellular components (C).
(Figure 6B). Most DEPs are involved in the cluster of were analyzed by immunostaining and Western blot.
metabolite interconversion enzymes (five proteins). The The levels of both CPT1B and ACAA2 increased in the
result is consistent with that in control group-specific hearts of HFD-fed mice (Figures 7 and 8). The result is
DEPs. In the KEGG pathway, 7 DEPs were distributed in consistent with the previous proteomic analysis.
seven pathways.
4. Discussion
3.7. Elevation of fatty acid metabolism-related Heart failure is the leading cause of MC [17] . Many
enzymes in the hearts of HFD-fed mice
complex molecular mechanisms are reportedly play a
Since the cluster of metabolite interconversion enzymes role in the pathogenic process of MC. Despite that, the
has the most DEPs (5 proteins) and fatty acid metabolism changes of proteome in MC remain unknown. In this
plays critical roles in the MC, protein levels of two fatty study, we used LC-MS/MS to observe the significant
acid metabolism-related DEPs, CPT1B and ACAA2, difference in proteome between standard diet-fed
Volume 1 Issue 2 (2022) 5 https://doi.org/10.36922/gtm.v1i2.137