Page 63 - GTM-2-3
P. 63
Global Translational Medicine Influence of ferroptosis in neurological diseases
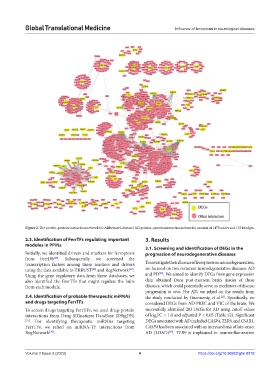
Figure 2. The protein–protein interaction network for Alzheimer’s disease (AD protein–protein interaction networks) consists of 1478 nodes and 13344 edges.
2.3. Identification of FerrTFs regulating important 3. Results
modules in PPINs
3.1. Screening and identification of DEGs in the
Initially, we identified drivers and markers for ferroptosis progression of neurodegenerative diseases
from FerrDb . Subsequently, we screened the
[28]
transcription factors among these markers and drivers To investigate the influence of ferroptosis on neurodegeneration,
using the data available in TRRUST and RegNetwork . we focused on two common neurodegenerative diseases: AD
[30]
[29]
[32]
Using the gene regulatory data from these databases, we and PD . We aimed to identify DEGs from gene expression
also identified the FerrTFs that might regulate the hubs data obtained from post-mortem brain tissues of these
from each module. diseases, which could potentially serve as mediators of disease
progression in vivo. For AD, we relied on the results from
2.4. Identification of probable therapeutic miRNAs the study conducted by Guennewig et al. . Specifically, we
[21]
and drugs targeting FerrTFs considered DEGs from AD-PREC and VIC of the brain. We
To screen drugs targeting FerrTFs, we used drug-protein successfully identified 283 DEGs for AD using cutoff values
interactions from Drug SIGnatures DataBase (DSigDB) of log FC > 1.0 and adjusted P < 0.05 (Table S2). Significant
2
[31] . For identifying therapeutic miRNAs targeting DEGs associated with AD included CASP4, TLR9, and C5AR1.
FerrTFs, we relied on miRNA-TF interactions from CASP4 has been associated with an increased risk of late-onset
RegNetwork . AD (LOAD) . TLR9 is implicated in neuroinflammation
[33]
[30]
Volume 2 Issue 3 (2023) 4 https://doi.org/10.36922/gtm.0318