Page 65 - GTM-2-3
P. 65
Global Translational Medicine Influence of ferroptosis in neurological diseases
A B
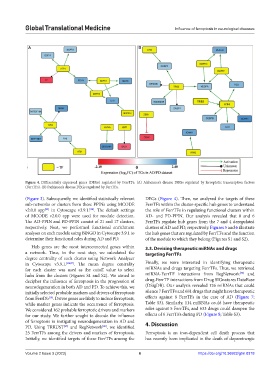
Figure 4. Differentially expressed genes (DEGs) regulated by FerrTFs. (A) Alzheimer’s disease DEGs regulated by ferroptotic transcription factors
(FerrTFs). (B) Parkinson’s disease DEGs regulated by FerrTFs.
(Figure 3). Subsequently, we identified statistically relevant DEGs (Figure 4). Then, we analyzed the targets of these
sub-networks or clusters from these PPINs using MCODE FerrTFs within the cluster-specific hub genes to understand
v2.0.0 app in Cytoscape v3.9.1. . The default settings the role of FerrTFs in regulating functional clusters within
[25]
[24]
of MCODE v2.0.0 app were used for module detection. AD- and PD-PPIN. Our analysis revealed that 8 and 6
The AD-PPIN and PD-PPIN consist of 21 and 17 clusters, FerrTFs regulate hub genes from the 7 and 4 deregulated
respectively. Next, we performed functional enrichment clusters of AD and PD, respectively. Figures 5 and 6 illustrate
analyses on each module using BiNGO in Cytoscape 3.9.1 to the hub genes that are regulated by FerrTFs and the function
determine their functional roles during AD and PD. of the module to which they belong (Figures S1 and S2).
Hub genes are the most interconnected genes within 3.3. Devising therapeutic miRNAs and drugs
a network. Thus, in the next step, we calculated the targeting FerrTFs
degree centrality of each cluster using Network Analyzer
in Cytoscape v3.9.1. [24,27] . The mean degree centrality Finally, we were interested in identifying therapeutic
for each cluster was used as the cutoff value to select miRNAs and drugs targeting FerrTFs. Thus, we retrieved
[30]
hubs from the clusters (Figures S1 and S2). We aimed to miRNA-FerrTF interactions from RegNetwork and
decipher the influence of ferroptosis in the progression of drug-FerrTF interactions from Drug SIGnatures DataBase
neurodegeneration in both AD and PD. To achieve this, we (DSigDB). Our analysis revealed 126 miRNAs that could
initially selected probable markers and drivers of ferroptosis silence 7 FerrTFs and 681 drugs that might have therapeutic
from FerrDb . Driver genes are likely to induce ferroptosis, effects against 8 FerrTFs in the case of AD (Figure 7;
[28]
while marker genes indicate the occurrence of ferroptosis. Table S3). Similarly, 114 miRNAs could have therapeutic
We considered 162 probable ferroptotic drivers and markers roles against 5 FerrTFs, and 633 drugs could dampen the
for our study. We further sought to decode the influence effects of 6 FerrTFs during PD (Figure 8; Table S3).
of ferroptosis in instigating neurodegeneration in AD and 4. Discussion
[29]
[30]
PD. Using TRRUST and RegNetwork , we identified
25 FerrTFs among the drivers and markers of ferroptosis. Ferroptosis is an iron-dependent cell death process that
Initially, we identified targets of these FerrTFs among the has recently been implicated in the death of dopaminergic
Volume 2 Issue 3 (2023) 6 https://doi.org/10.36922/gtm.0318