Page 67 - GTM-2-3
P. 67
Global Translational Medicine Influence of ferroptosis in neurological diseases
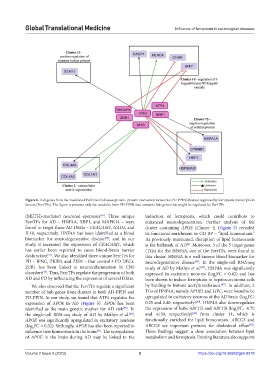
Figure 6. Hub genes from the modules of Parkinson’s disease protein–protein interaction networks (PD-PPIN) that are regulated by ferroptotic transcription
factors (FerrTFs). The figure represents only the modules from PD-PPIN that contains hub genes that might be regulated by FerrTFs.
(METH)-mediated neuronal apoptosis . Three unique induction of ferroptosis, which could contribute to
[55]
FerrTFs for AD – HNF4A, XBP1, and MAPK14 – were enhanced neurodegeneration. Further analysis of the
found to target three AD DEGs – CEACAM1, GAD1, and cluster containing APOE [Cluster 4] (Figure 5) revealed
IL18, respectively. HNF4A has been identified as a blood its functional enrichment in GO BP – “lipid homeostasis.”
biomarker for neurodegenerative disease , and in our As previously mentioned, disruption of lipid homeostasis
[56]
study, it increased the expression of CEACAM1, which is the hallmark of AD . Moreover, 3 of the 5 target genes
[8]
has earlier been reported to cause blood–brain barrier (TGs) for the HNF4A, one of the FerrTFs, were found in
dysfunction . We also identified three unique FerrTFs for this cluster. HNF4A is a well-known blood biomarker for
[57]
PD – IFNG, TRIB3, and ZEB1 – that control 4 PD DEGs. neurodegenerative disease . In the single-cell RNA-seq
[56]
ZEB1 has been linked to neuroinflammation in CNS study of AD by Mathys et al. , HNF4A was significantly
[60]
disorders . Thus, FerrTFs regulate the progression of both expressed in excitatory neurons (log FC = 0.42) and has
[58]
2
AD and PD by influencing the expression of several DEGs. been shown to induce ferroptosis in hepatocarcinoma cells
[47]
We also observed that the FerrTFs regulate a significant by binding to histone acetyltransferases. . In addition, 2
number of hub genes from clusters in both AD-PPIN and TGs of HNF4A, namely APOE1 and LIPC, were found to be
PD-PPIN. In our study, we found that ATF4 regulates the upregulated in excitatory neurons of the AD brain (log FC:
2
[60]
expression of APOE in AD (Figure 5). APOE has been 0.25 and 0.40, respectively) . HNF4A also downregulates
identified as the main genetic marker for AD risk . In the expression of hubs ABCG5 and ABCG8 (log FC: -0.70
[59]
2
[60]
the single-cell RNA-seq study of AD by Mathys et al. , and -0.30, respectively) from cluster 11, which is
[60]
APOE was significantly upregulated in excitatory neurons functionally enriched for lipid homeostasis. ABCG5 and
[62]
(log FC = 0.32). Strikingly, APOE has also been reported to ABCG8 are important proteins for cholesterol efflux .
2
influence iron homeostasis in the brain . The upregulation These findings suggest a close association between lipid
[61]
of APOE in the brain during AD may be linked to the metabolism and ferroptosis. Existing literature also supports
Volume 2 Issue 3 (2023) 8 https://doi.org/10.36922/gtm.0318