Page 71 - IJAMD-1-2
P. 71
International Journal of AI for
Materials and Design
Machine learning for gel fraction prediction
A B D
C
E
F G H
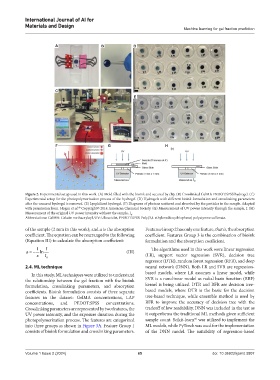
Figure 2. Experimental setup used in this work. (A) Mold filled with the bioink and secured by clip. (B) Crosslinked GelMA-PEDOT:SPSS hydrogel. (C)
Experimental setup for the photopolymerization process of the hydrogel. (D) Hydrogels with different bioink formulation and crosslinking parameters
after the uncured hydrogel is removed. (E) Lyophilized hydrogel. (F) Diagrams of photons scattered and absorbed by the particles in the sample. Adapted
with permission from. Hogan et al. Copyright© 2014 American Chemical Society. (G) Measurement of UV power intensity through the sample, I. (H)
49
Measurement of the original UV power intensity without the sample, I .
0
Abbreviations: GelMA: Gelatin methacryloyl, UV: Ultraviolet, PEDOT:SPSS: Poly(3,4-ethylenedioxythiophene) polystyrene sulfonate.
of the sample (2 mm in this work); and a is the absorption Features Group 2 has only one feature, that is, the absorption
coefficient. The equation can be rearranged to the following coefficient. Features Group 3 is the combination of bioink
(Equation III) to calculate the absorption coefficient: formulation and the absorption coefficient.
1 I The algorithms used in this work were linear regression
a ln (III)
x I 0 (LR), support vector regression (SVR), decision tree
regressor (DTR), random forest regression (RFR), and deep
2.4. ML technique neural network (DNN). Both LR and SVR are regression-
In this study, ML techniques were utilized to understand based models, where LR assumes a linear model, while
the relationship between the gel fraction with the bioink SVR is a non-linear model as radial basis function (RBF)
formulation, crosslinking parameters, and absorption kernel is being utilized. DTR and RFR are decision tree-
coefficients. Bioink formulation consists of three separate based models, where DTR is the basic for the decision
features in the dataset: GelMA concentrations, LAP tree-based technique, while ensemble method is used by
concentrations, and PEDOT:SPSS concentrations. RFR to improve the accuracy of decision tree with the
Crosslinking parameters are represented by two features, the tradeoff of low readability. DNN was included in the test as
UV power intensity, and the exposure duration during the it outperforms the traditional ML methods given sufficient
50
photopolymerization process. The features are categorized sample count. Scikit-learn was utilized to implement the
into three groups as shown in Figure 3A. Feature Group 1 ML models, while PyTorch was used for the implementation
consists of bioink formulation and crosslinking parameters. of the DNN model. The suitability of regression-based
Volume 1 Issue 2 (2024) 65 doi: 10.36922/ijamd.3807