Page 281 - IJB-10-2
P. 281
International Journal of Bioprinting Microfluidic spinning for neural models
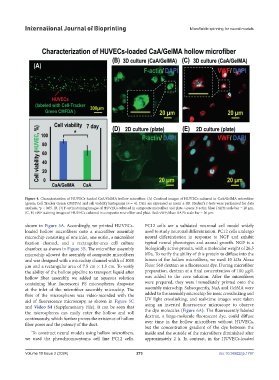
Figure 4. Characterization of HUVECs-loaded CaA/GelMA hollow microfiber. (A) Confocal images of HUVECs cultured in CaA/GelMA microfiber
(green: Cell Tracker Green CMFDA) and cell viability histograms (n = 4). Data are expressed as mean ± SD. Student’s t-tests were performed for data
analysis. *p < 0.05. (B, D) F-actin staining images of HUVECs cultured in composite microfiber and plate. Green: F-actin; blue: DAPI; scale bar = 20 µm.
(C, E) vWF staining images of HUVECs cultured in composite microfiber and plate. Red: vWF; blue: DAPI; scale bar = 20 µm.
shown in Figure 5A. Accordingly, we printed HUVECs- PC12 cells are a validated neuronal cell model widely
loaded hollow microfibers onto a microfiber assembly used to study neuronal differentiation. PC12 cells undergo
microchip consisting of one inlet, one outlet, a microfiber neural differentiation in response to NGF and exhibit
fixation channel, and a rectangular-area cell culture typical neural phenotypes and axonal growth. NGF is a
chamber, as shown in Figure 5B. The microfiber assembly biologically active protein, with a molecular weight of 26.5
microchip allowed the assembly of composite microfibers kDa. To verify the ability of this protein to diffuse into the
and was designed with a microchip channel width of 1000 lumen of the hollow microfibers, we used 10 kDa Alexa
μm and a rectangular area of 7.5 cm × 1.5 cm. To verify Fluor 568 dextran as a fluorescent dye. During microfiber
the ability of the hollow pipeline to transport liquid after preparation, dextran at a final concentration of 100 μg/L
hollow fiber assembly, we added an aqueous solution was added to the core solution. After the microfibers
containing blue fluorescent PS microspheres dropwise were prepared, they were immediately printed onto the
at the inlet of the microfiber assembly microchip. The assembly microchip. Subsequently, NaA and GelMA were
flow of the microspheres was video-recorded with the added to the assembly microchip for ionic crosslinking and
aid of fluorescence microscopy, as shown in Figure 5C UV light crosslinking, and real-time images were taken
and Video S4 (Supplementary File). It can be seen that using an inverted fluorescence microscope to observe
the microspheres can easily enter the hollow and roll the dye molecules (Figure 6A). The fluorescently labeled
continuously, which further proves the existence of hollow dextran, a large-molecule fluorescent dye, could diffuse
fiber pores and the patency of the duct. over time in the hollow microfibers without HUVECs,
but the concentration gradient of the dye between the
To construct neural models using hollow microfibers, inside and the outside of the microfibers diminished after
we used the pheochromocytoma cell line PC12 cells. approximately 2 h. In contrast, in the HUVECs-loaded
Volume 10 Issue 2 (2024) 273 doi: 10.36922/ijb.1797