Page 13 - IJB-10-4
P. 13
International Journal of Bioprinting PAI for 3D bioprinted constructs
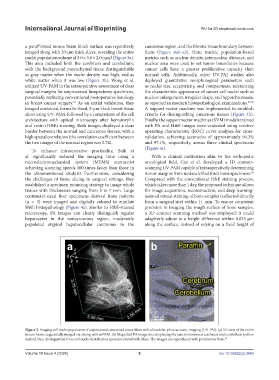
a paraffinized mouse brain block surface was repetitively cancerous region, and the fibrotic tissue boundary between
imaged along with 20-μm thick slices, revealing the entire them (Figure 4a1–a3). Here, nucleic population-based
nuclei population volume of 3.0 × 3.8 × 2.0 mm3 (Figure 3a). metrics such as nuclear density, internuclear distance, and
This area included both the cerebrum and cerebellum, nuclear area were used to set tumor boundaries because
with the background parenchymal tissue distinguishable cancer cells have a greater proliferative capacity than
as gray matter when the nuclei density was high, and as normal cells. Additionally, other UV-PAI studies also
white matter when it was low (Figure 3b). Wong et al. deployed quantitative morphological parameters such
utilized UV-PAM in the intraoperative assessment of clear as nuclei size, eccentricity, and compactness, referencing
surgical margins for unprocessed lumpectomy specimens, the characteristic appearance of cancer cell nuclei such as
potentially replacing conventional postoperative histology nuclear enlargement, irregular shape, and hyperchromasia,
in breast cancer surgery. As an initial validation, they as reported in modern histopathological examinations. 65,66
63
imaged unstained, formalin-fixed, 5-μm-thick breast tissue A support vector machine was implemented to establish
slices using UV-PAM, followed by a comparison of the cell criteria for distinguishing cancerous tissues (Figure 4b).
architecture with optical microscopy after hematoxylin Finally, the support vector machines (SVM) models trained
and eosin (H&E) staining. Both images displayed a clear with PA and H&E images were evaluated using receiver
border between the normal and cancerous tissues, with a operating characteristic (ROC) curve analyses for cross-
high spatial correlation (the correlation coefficient between validation, achieving accuracies of approximately 94.3%
the two images of the normal region was 0.74). and 97.1%, respectively, across three clinical specimens
(Figure 4c).
To enhance intraoperative practicality, Baik et
al. significantly reduced the imaging time using a With a clinical motivation akin to the orthopedic
microelectromechanical system (MEMS) scanner,64 oncological field, Cao et al. developed a 3D contour-
achieving scanning speeds 100 times faster than those in scanning UV-PAM capable of intraoperatively determining
the aforementioned study.62 Furthermore, considering tumor margins from undecalcified thick bone specimens.
66
the challenges of tissue slicing in surgical settings, they Compared with the conventional H&E staining process,
established a specimen mounting strategy to image whole which takes more than 1 day, the proposed technique allows
tissues with thicknesses ranging from 3 to 7 mm. Large for image acquisition, reconstruction, and deep learning-
centimeter-sized liver specimens derived from patients assisted virtual staining of bone samples (collected directly
(n = 3) were imaged and digitally colored to emulate from a surgical site) within 11 min. To ensure consistent
H&E histopathology (Figure 4a). Similar to H&E-stained precision in imaging the rough surface of bone samples,
microscopy, PA images can clearly distinguish regular a 3D contour scanning method was employed; it could
hepatocytes in the noncancerous region, moderately adaptively adjust to a height difference within 0.625 μm
populated atypical hepatocellular carcinoma in the along the surface, instead of relying on a fixed height of
Figure 3. Imaging cell nuclei population of unprocessed, unstained tissue slices with ultraviolet-photoacoustic imaging (UV-PAI). (a) 3D view of the entire
mouse brain, sequentially imaged via slicing with mPAM. (b) Magnified PA image slice displaying the junction between cerebrum and cerebellum (yellow
dashed line), distinguished from cell nuclei distribution (pseudocolored with blue). The images are reproduced with permission from. 62
Volume 10 Issue 4 (2024) 5 doi: 10.36922/ijb.3448