Page 45 - IJB-5-1
P. 45
Cheptsov VS, et al.
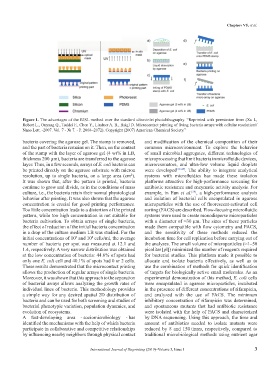
Figure 1. The advantages of the RISL method over the standard ultraviolet photolithography. “Reprinted with permission from (Xu L,
Robert L., Ouyang Q., Taddei F., Chen Y., Lindner A. B., Baigl D. Microcontact printing of living bacteria arrays with cellular resolution//
Nano Lett. -2007. Vol. 7 - № 7. - P. 2068–2072). Copyright (2007) American Chemical Society.”
bacteria covering the agarose gel. The stamp is removed, and modification of the chemical composition of their
and the part of bacteria remains on it. Then, on the contact common microenvironment. To explore the behavior
of the stamp with the layer of agarose gel (4 wt% in LB, of small microbial aggregates, different technologies of
thickness 200 µm), bacteria are transferred to the agarose microprocessing that limit bacteria in microfluidic devices,
layer. Thus, in a few seconds, arrays of E. coli bacteria can microresonators, and ultra-low volume liquid droplets
be printed directly on the agarose substrate with micron were developed [39-45] . The ability to integrate analytical
resolution, up to single bacteria, on a large area (cm ). systems with microfluidics has made these isolation
2
It was shown that, after the pattern is printed, bacteria platforms attractive for high-performance screening for
continue to grow and divide, as in the conditions of mass antibiotic resistance and enzymatic activity analysis. For
culture, i.e., the bacteria retain their normal physiological example, in Eun et al. , a high-performance analysis
[39]
behavior after printing. It was also shown that the agarose and isolation of bacterial cells encapsulated in agarose
concentration is crucial for good printing performance. microparticles with the use of fluorescent-activated cell
Too little concentration leads to a distortion of the printed sorting (FACS) are described. Flow-focusing microfluidic
pattern, while too high concentration is not suitable for systems were used to create monodisperse microparticles
bacteria cultivation. To obtain arrays of single bacteria, with a diameter of ≈30 µm. The sizes of these particles
the effect of reduction of the initial bacteria concentration made them compatible with flow cytometry and FACS,
in a drop of the culture medium LB was studied. For the and the sensitivity of these methods reduced the
initial concentrations of 10 and 10 cells/ml, the average incubation time for cell replication before carrying out of
8
9
number of bacteria per spot was measured at 12.1 and the analyses. The small volume of microparticles (≈1–50
1.4, respectively. A very narrow distribution was obtained picoliter [pl]) minimized the number of reagents required
at the low concentration of bacteria: 44.6% of spots had for bacterial studies. This platform made it possible to
only one E. coli cell and 40.1% of spots had 0 or 2 cells. allocate and isolate bacteria effectively, as well as to
These results demonstrated that the microcontact printing use the combination of methods for quick identification
allows the production of regular arrays of single bacteria. of targets for biologically active small molecules. As an
Moreover, it was shown that this approach to the separation experimental demonstration of this method, E. coli cells
of bacterial arrays allows analyzing the growth rates of were encapsulated in agarose microparticles, incubated
individual lines of bacteria. This methodology provides in the presence of different concentrations of rifampicin,
a simple way for any desired spatial 2D distribution of and analyzed with the use of FACS. The minimum
bacteria and can be used for both screening and studies of inhibitory concentration of rifampicin was determined,
bacterial phenotypic variation, population dynamics, and and spontaneous mutants that had antibiotic resistance
evolution of ecosystems. were isolated with the help of FACS and characterized
A fast-developing area - sociomicrobiology - has by DNA sequencing. Using this approach, the time and
identified the mechanisms with the help of which bacteria amount of antibiotics needed to isolate mutants were
participate in collaborative and competitive relationships reduced by 8 and 150 times, respectively, compared to
by influencing nearby neighbors through physical contact traditional microbiological methods using nutrient agar
International Journal of Bioprinting (2019)–Volume 5, Issue 1 3