Page 545 - IJB-10-6
P. 545
International Journal of Bioprinting Design and property of PLPG/PDLA scaffold
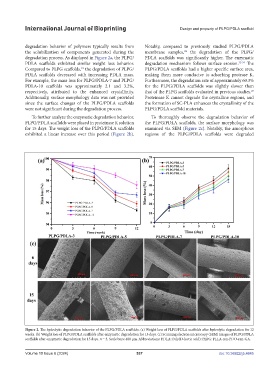
degradation behavior of polymers typically results from Notably, compared to previously studied PLPG/PDLA
the solubilization of components generated during the membrane samples, the degradation of the PLPG/
24
degradation process. As displayed in Figure 2a, the PLPG/ PDLA scaffolds was significantly higher. The enzymatic
PDLA scaffolds exhibited similar weight loss behavior. degradation mechanism follows surface erosion. 32-34 The
Compared to PLPG scaffolds, the degradation of PLPG/ PLPG/PDLA scaffolds had a higher specific surface area,
18
PDLA scaffolds decreased with increasing PDLA mass. making them more conducive to adsorbing protease K.
For example, the mass loss for PLPG/PDLA-7 and PLPG/ Furthermore, the degradation rate of approximately 60.3%
PDLA-10 scaffolds was approximately 2.1 and 3.2%, for the PLPG/PDLA scaffolds was slightly slower than
respectively, attributed to the enhanced crystallinity. that of the PLPG scaffolds evaluated in previous studies.
29
Additionally, surface morphology data was not provided Proteinase K cannot degrade the crystalline regions, and
since the surface changes of the PLPG/PDLA scaffolds the formation of SC-PLA enhances the crystallinity of the
were not significant during the degradation process. PLPG/PDLA scaffold materials.
To further analyze the enzymatic degradation behavior, To thoroughly observe the degradation behavior of
PLPG/PDLA scaffolds were placed in proteinase K solution the PLPG/PDLA scaffolds, the surface morphology was
for 15 days. The weight loss of the PLPG/PDLA scaffolds examined via SEM (Figure 2c). Notably, the amorphous
exhibited a linear increase over this period (Figure 2b). regions of the PLPG/PDLA scaffolds were degraded
Figure 2. The hydrolytic degradation behavior of the PLPG/PDLA scaffolds. (a) Weight loss of PLPG/PDLA scaffolds after hydrolytic degradation for 12
weeks. (b) Weight loss of PLPG/PDLA scaffolds after enzymatic degradation for 15 days. (c) Scanning electron microscopy (SEM) images of PLPG/PDLA
scaffolds after enzymatic degradation for 15 days. n = 3. Scale bars: 400 μm. Abbreviations: PDLA: Poly(D-lactic acid); PLPG: PLLA-ran-PDO-ran-GA.
Volume 10 Issue 6 (2024) 537 doi: 10.36922/ijb.4645