Page 118 - ITPS-7-1
P. 118
INNOSC Theranostics and
Pharmacological Sciences Repurposed Drugs as inhibitors of Pfmrk
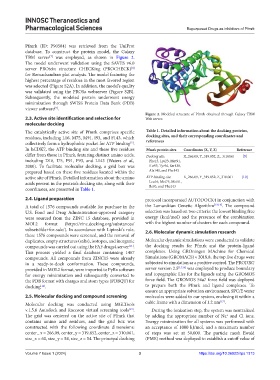
Pfmrk (ID: P90584) was retrieved from the UniProt
database. To construct the protein model, the Galaxy
TBM server was employed, as shown in Figure 2.
[7]
The model underwent validation using the SAVES v6.0
[8]
server PROtein structure CHECKing (PROCHECK)
for Ramachandran plot analysis. The model featuring the
highest percentage of residues in the most favored region
was selected (Figure S2A). In addition, the model’s quality
was validated using the PROSa webserver (Figure S2B).
Subsequently, the modeled protein underwent energy
minimization through SWISS Protein Data Bank (PDB)
viewer software .
[9]
Figure 2. Modeled structure of Pfmrk obtained through Galaxy TBM
2.3. Active site identification and selection for Web server.
molecular docking
The catalytically active site of Pfmrk comprises specific Table 1. Detailed information about the docking proteins,
residues, including L16, M75, M91, I93, and F143, which docking sites, and their corresponding coordinates and
collectively form a hydrophobic pocket for ATP binding . references
[5]
In hCDK7, the ATP binding site and these five residues Pfmrk protein sites Coordinates (X, Y, Z) Reference
differ from those in Pfmrk, featuring distinct amino acids, Docking site: X_266.09, Y_319.852, Z_ 310.061 [5]
including D16, I75, F91, F93, and L143 (Waters et al., Phe15, Lys23, Met91,
2000). To facilitate molecular docking, a grid box was ILe93, Tyr96, Ser138,
prepared based on these five residues located within the Ala140, and Phe143
active site of Pfmrk. Detailed information about the amino ATP binding site: X_266.09, Y_319.852, Z_310.061 [10]
acids present in the protein’s docking site, along with their Leu16, Met75, Met91,
coordinates, are presented in Table 1. Ile93, and Phe143
2.4. Ligand preparation protocol incorporated AUTODOCK4 in conjunction with
A total of 1576 compounds available for purchase in the the Lamarckian Genetic Algorithm [13,14] . The compound
U.S. Food and Drug Administration-approved category selection was based on two criteria: the lowest binding free
were sourced from the ZINC 15 database, provided in energy (kcal/mol) and the presence of the combination
MOL2 format (https://zinc.docking.org/substances/ with the highest number of clusters for each compound.
subsets/fda+for-sale/). In accordance with Lipinski’s rule, 2.6. Molecular dynamic simulation research
these 1576 compounds were screened, and the removal of
duplicates, empty structures (salts), isotopes, and inorganic Molecular dynamic simulations were conducted to validate
compounds was carried out using the FAF drugs4 server . the docking results for Pfmrk and the protein-ligand
[11]
This process yielded a final database containing 1467 complexes. Using GROningen MAchine for Chemical
compounds. All compounds from ZINC15 were already Simulations (GROMACS) v 2018.8, the top five drugs were
in a ready-to-dock conformation. These compounds, subjected to simulation as a positive control. The PRODRG
provided in MOL2 format, were imported to PyRx software server version 2.5 [15,16] was employed to produce boundary
for energy minimization and subsequently converted to and topographic files for the ligands using the GROMOS
the PDB format with charges and atom types (PDBQT) for force field. The GROMOS 54a7 force field was deployed
docking . to prepare both the Pfmrk and ligand complexes. To
[12]
ensure an appropriate solvation environment, SPC/E water
2.5. Molecular docking and compound screening molecules were added to our system, enclosing it within a
[17]
Molecular docking was conducted using MGLTools cubic frame with a dimension of 1.2 nm .
v.1.5.6 Autodock and Raccoon virtual screening tools . During the ionization step, the system was neutralized
[13]
The grid was centered on the active site of Pfmrk that by adding the appropriate number of Na and Cl ions.
-
+
contains amino acid residues, and the grid box was Energy minimization for all systems was performed with
constructed with the following coordinate dimensions: an acceptance of 1000 kJ/mol, and a maximum number
center_ x = 266.09, center_y = 319.852, center_z = 310.061, of steps was set at 50,000. The particle mesh Ewald
size_x = 60, size_y = 54, size_z = 54. The principal docking (PME) method was deployed to establish a cutoff value of
Volume 7 Issue 1 (2024) 3 https://doi.org/10.36922/itps.1313