Page 46 - ITPS-8-2
P. 46
INNOSC Theranostics and
Pharmacological Sciences MDD biomarkers: Clinical implications
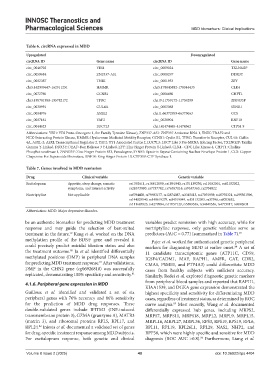
Table 6. circRNA expressed in MDD
Upregulated Downregulated
circRNA ID Gene name circRNA ID Gene name
circ_0046702 YES1 circ_0009024 TXLNG2P
circ_0059684 ZNF337-AS1 circ_0008297 DDX3Y
circ_0002387 TNIK circ_0001953 ZFY
chr5:162909647-162911251 HMMR chr5:178043882-178044435 CLK4
circ_0072760 CCNB1 circ_0006660 CHPT1
chr3:195781950-195782172 TFRC chr19:11759172-11759299 ZNF833P
circ_0030991 CUL4A circ_0003068 SYNE1
circ_0004976 ASXL2 chr11:66372959-66373063 CCS
circ_0007161 YAF2 circ_0028904 RNF10
circ_0044623 LUC7L3 chr1:41474465-41474562 CTPS1 9
Abbreviations: YES1: YES Proto-Oncogene 1, Src Family Tyrosine Kinase), ZNF337-AS1: ZNF337 Antisense RNA 1, TNIK: TRAF2 and
NCK-Interacting Protein Kinase, HMMR: Hyaluronan Mediated Motility Receptor, CCNB1: Cyclin B1, TFRC: Transferrin Receptor, CUL4A: Cullin
4A, ASXL-2: ASXL Transcriptional Regulator 2, YAF2: YY1 Associated Factor 2, LUC7L3: LUC7 Like 3 Pre-MRNA Splicing Factor, TXLNG2P: Taxilin
Gamma Y-Linked, DDX3Y: DEAD-Box Helicase 3 Y-Linked, ZFY: Zinc Finger Protein Y-Linked, CLK4 - CDC Like Kinase 4, CHPT1: Choline
Phosphotransferase 1, ZNF833P: Zinc Finger Protein 833, Pseudogene, SYNE1: Spectrin Repeat Containing Nuclear Envelope Protein 1, CCS: Copper
Chaperone For Superoxide Dismutase, RNF10: Ring Finger Protein 10, CTPS19: CTP Synthase 1.
Table 7. Genes involved in MDD remission
Drug Clinical variable Genetic variable
Escitalopram Appetite, sleep change, somatic rs1392611, rs10812099, rs1891943, rs151139256, rs11002001, rs62182022,
symptoms, and interest activity rs28373080, rs7757702, rs76557116, rs9557363, rs2704022
Nortriptyline Not applicable rs6794400, rs79693177, rs12874087, rs2345113, rs17091959, rs10792321, rs199561596,
rs144829540, rs149619279, rs34319049, rs151132095, rs37596, rs8053632,
rs111685823, rs4279984, rs17057129, rs5889536, rs34841556, rs4773117, rs8082631
Abbreviation: MDD: Major depressive disorder.
be an authentic biomarker for predicting MDD treatment variables predict remission with high accuracy, while for
response and may guide the selection of best-suited nortriptyline response, only genetic variables serve as
79
treatment in the future. Kang et al. worked on the DNA predictors (AUC = 0.77) (summarized in Table 7). 83
methylation profile of the BDNF gene and revealed it Pajer et al. worked for authenticated genetic peripheral
could precisely predict suicidal ideation status and also markers for diagnosing MDD at earlier onset. A set of
84
the treatment outcome. Ju et al. identified differentially 11 candidate transcriptomic genes (ATP11C, CD59,
80
methylated positions (DMP) in peripheral DNA samples IGSF4/CADM1, MAF, RAPH1, AMFR, CAT, CDR2,
for predicting MDD treatment response. After validations, CMAS, PSME1, and PTP4A3) could differentiate MDD
81
DMP in the CHN2 gene (cg06926818) was successfully cases from healthy subjects with sufficient accuracy.
replicated, demonstrating 100% specificity and sensitivity. 81 Similarly, Redei et al. explored diagnostic genetic markers
4.1.6. Peripheral gene expression in MDD from peripheral blood samples and reported that RAPH1,
KIAA1539, and DGKA gene expression demonstrated the
Guilloux et al. identified and validated a set of six highest specificity and sensitivity for differentiating MDD
peripheral genes with 76% accuracy and 86% sensitivity cases, regardless of treatment status, as determined by ROC
for the prediction of MDD drug responses. These curve analysis. Most recently, Wang et al. documented
85
double-validated genes include IFITM3 (INF-induced differentially expressed hub genes, including MRPS2,
transmembrane protein 3), GZMA (granzyme A), MATR3 MRPS7, MRPS11, MRPS18, MRPL2, MRPL9, MRPL15,
(matrin 3), and ribosomal proteins RPL5, RPL17, and MRPL16, MRPL27, MRPL36, RPS3, RPS19, RPS19, RPL6,
RPL24. Iniesta et al. documented a validated set of genes RPL11, RPL19, RPL26L1, RPL29, NAS2, NHP2, and
82
for drug-specific treatment response among MDD subjects. RPP38, which were highly specific and sensitive for MDD
For escitalopram response, both genetic and clinical diagnosis (ROC AUC >0.8). Furthermore, Liang et al.
86
Volume 8 Issue 2 (2025) 40 doi: 10.36922/itps.4404