Page 63 - TD-3-3
P. 63
Tumor Discovery Pyroptosis-related genes in breast cancer progression
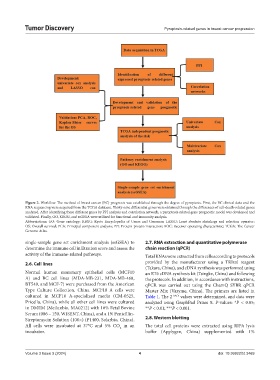
Figure 2. Workflow. The method of breast cancer (BC) prognosis was established through the degree of pyroptosis. First, the BC clinical data and the
RNA sequencing were acquired from the TCGA database. Thirty-nine differential genes were obtained through the differences of cell-death-related genes
analyzed. After identifying these different genes by PPI analysis and correlation network, a pyroptosis-related gene prognostic model was developed and
validated. Finally, GO, KEGG, and ssGESA were utilized for functional and immunity analysis.
Abbreviations: GO: Gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; LASSO: Least absolute shrinkage and selection operator;
OS: Overall survival; PCA: Principal component analysis; PPI: Protein-protein interaction; ROC: Receiver operating characteristic; TCGA: The Cancer
Genome Atlas.
single-sample gene set enrichment analysis (ssGSEA) to 2.7. RNA extraction and quantitative polymerase
determine the immune cell infiltration score and assess the chain reaction (qPCR)
activity of the immune-related pathways. Total RNAs were extracted from cells according to protocols
2.6. Cell lines provided by the manufacturer using a TRIzol reagent
(Takara, China), and cDNA synthesis was performed using
Normal human mammary epithelial cells (MCF10 an RT6 cDNA synthesis kit (Tsingke, China) and following
A) and BC cell lines (MDA-MB-231, MDA-MB-468, the protocols. In addition, in accordance with instructions,
BT549, and MCF-7) were purchased from the American qPCR was carried out using the ChamQ SYBR qPCR
Type Culture Collection, China. MCF10 A cells were Master Mix (Vazyme, China). The primers are listed in
cultured in MCF10 A-specialized media (CM-0525, Table 1. The 2 -ΔΔCt values were determined, and data were
Pricella, China), while all other cell lines were cultured analyzed using GraphPad Prism 8. P-values: *P < 0.05;
in DMEM (Meilunbio, MA0212) with 10% Fetal Bovine **P < 0.01; ***P < 0.001.
Serum (086 – 150, WISENT, China), and a 1% Penicillin-
Streptomycin Solution (100×) (P1400, Solarbio, China). 2.8. Western blotting
All cells were incubated at 37°C and 5% CO in an The total cell proteins were extracted using RIPA lysis
2
incubator. buffer (Applygen, China) supplemented with 1%
Volume 3 Issue 3 (2024) 4 doi: 10.36922/td.3469