Page 106 - TD-3-4
P. 106
Tumor Discovery Bioinformatics insights into CCL2 mutations
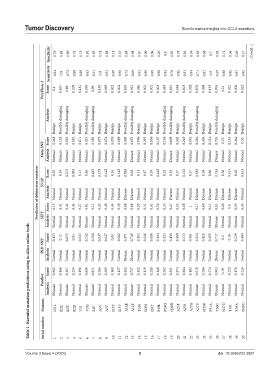
Specificity 0.79 0.84 0.89 0.74 0.73 0.92 0.95 0.71 0.68 0.18 0.35 0.88 0.91 0.67 0.09 0.09 0.26 0.8 0.85 0.78 0.83 0.59 0.89 0.68 0.7 0.93 0.74 0.56 0.45 0.57 (Cont’d...)
Sensitivity 0.84 0.8 0.72 0.88 0.89 0.63 0.51 0.9 0.91 0.99 0.98 0.73 0.68 0.91 0.99 0.99 0.98 0.83 0.78 0.85 0.81 0.94 0.71 0.91 0.9 0.59 0.88 0.95 0.97 0.95
PolyPhen-2 Score 0.4 0.637 0.86 0.209 0.181 0.959 0.99 0.139 0.098 0.002 0.004 0.839 0.921 0.086 0.001 0.001 0.003 0.449 0.691 0.348 0.603 0.028 0.876 0.098 0.119 0.976 0.21 0.021 0.006 0.022
Analysis Possibly damaging Possibly damaging Possibly damaging Possibly damaging Possibly damaging Possibly damaging Possibly damaging Possibly damaging Possibly damaging Possibly damaging Possibly damaging
Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign Benign
Meta-SNP Score Analysis 0.248 Normal 0.128 Normal 0.085 Normal 0.082 Normal 0.071 Normal 0.422 Normal 0.383 Normal 0.075 Normal 0.074 Normal 0.076 Normal 0.089 Normal 0.498 Normal 0.653 Disease 0.096 Normal 0.293 Normal 0.056 Normal 0.167 Normal 0.136 Normal 0.609 Disease 0.302 Normal 0.249 Normal 0.092 Normal 0.484 Normal 0.304 Normal 0.314 Normal 0.795 Disease 0.25 Normal 0.145 Normal 0.244 N
Prediction of deleterious mutation SNAP Score Analysis 0.25 Normal 0.15 0.34 Normal 0.41 0.275 Normal 0.42 0.095 Normal 0.44 0.13 Normal 0.27 0.46 Normal 0.01 0.485 Normal 0.11 0.175 Normal 0.42 0.145 Normal 0.48 0.16 Normal 0.44 0.145 Normal 0.38 0.585 Disease 0.08 0.62 Disease 0.08 0.13 Normal 0.55 0.37 Normal 0.31 0.19 Normal 0.41 0.245 Normal 0.76 0.32 Normal 0.35 0.55 Disease 0.47 0.37 Nor
Analysis SIFT Score Normal Normal Normal Normal Normal Disease Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Disease Normal Normal Normal Normal
Table 1. Harmful mutation predictions using in silico online tools
PhD-SNP Score 0.153 0.12 0.075 0.05 0.033 0.242 0.208 0.037 0.047 0.05 0.082 0.571 0.749 0.103 0.346 0.088 0.134 0.123 0.498 0.369 0.213 0.081 0.324 0.425 0.355 0.717 0.2 0.18 0.229 0.384
Analysis Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Disease Disease Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Normal Disease Normal Normal Normal Normal
Score 0.982 0.466 0.567 0.255 0.196 0.696 0.871 0.266 0.269 0.169 0.127 0.482 0.727 0.122 0.119 0.285 0.148 0.262 0.351 0.374 0.384 0.183 0.619 0.286 0.274 0.903 0.19 0.222 0.178 0.226
Panther
Analysis Disease Normal Disease Normal Normal Disease Disease Normal Normal Normal Normal Normal Disease Normal Normal Normal Normal Normal Normal Normal Normal Normal Disease Normal Normal Disease Normal Normal Normal Normal
Mutants M1L K2E K2T K2R V3I V3G S4F A5V A5T I13T I13V A14E A15P T16S I18N I18T P19L P19Q Q20R A23T A23S A27G A27V N29K P31A Y36C N37D T39I T39A N40D
Serial number 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30
Volume 3 Issue 4 (2024) 8 doi: 10.36922/td.3891