Page 111 - TD-3-4
P. 111
Tumor Discovery Bioinformatics insights into CCL2 mutations
A
B
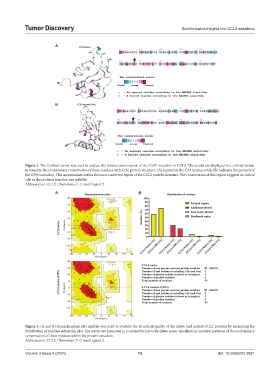
Figure 2. The ConSurf server was used to analyze the residue conservation of the C59G mutation in CCL2. The results are displayed in a colored format
to visualize the evolutionary conservation of these residues within the protein structure. (A) represents the C59 residue, while (B) indicates the position of
the C59G mutation. This mutation lies within the most conserved region of the CCL2 protein structure. The conservation of this region suggests its critical
role in the protein’s function and stability.
Abbreviation: CCL2: Chemokine C-C motif ligand 2.
A B
Figure 3. (A and B) Ramachandran plot analysis was used to evaluate the structural quality of the native and mutant CCL2 proteins by examining the
distribution of residues within the plot. The results are presented in a colored format to facilitate easier visualization and interpretation of the evolutionary
conservation of these residues within the protein structure.
Abbreviation: CCL2: Chemokine C-C motif ligand 2.
Volume 3 Issue 4 (2024) 13 doi: 10.36922/td.3891