Page 112 - TD-3-4
P. 112
Tumor Discovery Bioinformatics insights into CCL2 mutations
introduced considerable conformational changes that flexibility and structural alterations compared to the native
required more time to reach equilibrium (Figure 4A). CCL2 protein (Figure 5A and B).
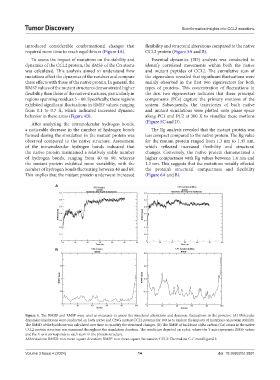
To assess the impact of mutations on the stability and Essential dynamics (ED) analysis was conducted to
dynamics of the CCL2 protein, the RMSF of the Cα atoms identify correlated movements within both the native
was calculated. This analysis aimed to understand how and mutant peptides of CCL2. The cumulative sum of
mutations affect the dynamics of the residues and compare the eigenvalues revealed that significant fluctuations were
these effects with those of the native protein. In general, the mainly observed in the first two eigenvectors for both
RMSF values of the mutant structures demonstrated higher types of proteins. This concentration of fluctuations in
flexibility than those of the native structures, particularly in the first two eigenvectors indicates that these principal
regions spanning residues 5 – 80. Specifically, these regions components (PCs) capture the primary motions of the
exhibited significant fluctuations in RMSF values, ranging system. Subsequently, the trajectories of both native
from 0.1 to 0.7 Å, which indicated increased dynamic and mutant simulations were plotted onto phase space
behavior in these areas (Figure 4B). along PC1 and PC2 at 300 K to visualize these motions
After analyzing the intramolecular hydrogen bonds, (Figure 5C and D).
a noticeable decrease in the number of hydrogen bonds The Rg analysis revealed that the mutant protein was
formed during the simulation in the mutant protein was less compact compared to the native protein. The Rg value
observed compared to the native structure. Assessment for the mutant protein ranged from 1.3 nm to 1.45 nm,
of the intramolecular hydrogen bonds indicated that which reflected increased flexibility and structural
the native protein maintained a relatively stable number changes. Conversely, the native protein demonstrated a
of hydrogen bonds, ranging from 40 to 80, whereas higher compactness with Rg values between 1.4 nm and
the mutant protein exhibited more variability, with the 1.3 nm. This suggests that the mutations notably affected
number of hydrogen bonds fluctuating between 40 and 69. the protein’s structural compactness and flexibility
This implies that the mutant protein underwent increased (Figure 6A and B).
A
B
Figure 4. The RMSD and RMSF were used as measures to assess the structural alterations and dynamic fluctuations in the proteins. (A) Molecular
dynamics simulations were conducted on both native and C59G mutant CCL2 proteins for 100 ns to explore the impacts of mutations on protein stability.
The RMSD of the backbone was calculated over time to quantify the structural changes. (B) The RMSF of backbone alpha carbon (Cα) atoms in the native
CCL2 protein structure was examined throughout the simulation duration. The results are depicted on a plot, where the Y-axis represents RMSF values
and the X-axis corresponds to each atom in the protein structure.
Abbreviations: RMSD: root mean square deviation; RMSF: root mean square fluctuation; CCL2: Chemokine C-C motif ligand 2.
Volume 3 Issue 4 (2024) 14 doi: 10.36922/td.3891