Page 113 - TD-3-4
P. 113
Tumor Discovery Bioinformatics insights into CCL2 mutations
A B
C D
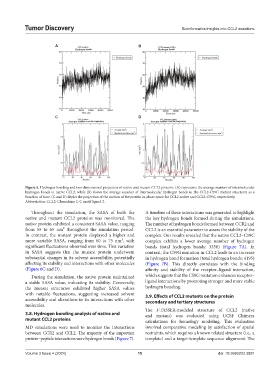
Figure 5. Hydrogen bonding and two-dimensional projection of native and mutant CCL2 proteins. (A) represents the average number of intermolecular
hydrogen bonds in native CCL2, while (B) shows the average number of intermolecular hydrogen bonds in the CCL2-C59G mutant structures as a
function of time. (C and D) depict the projection of the motion of the protein in phase space for CCL2-native and CCL2-C59G, respectively.
Abbreviation: CCL2: Chemokine C-C motif ligand 2.
Throughout the simulation, the SASA of both the A timeline of these interactions was generated to highlight
native and mutant CCL2 proteins was monitored. The the key hydrogen bonds formed during the simulations.
native protein exhibited a consistent SASA value, ranging The number of hydrogen bonds formed between CCR2 and
from 55 to 65 nm² throughout the simulation period. CCL2 is an essential parameter to assess the stability of the
In contrast, the mutant protein displayed a higher and complex. Our results revealed that the native CCL2–C59C
more variable SASA, ranging from 60 to 75 nm², with complex exhibits a lower average number of hydrogen
significant fluctuations observed over time. This variation bonds (total hydrogen bonds: 3350) (Figure 7A). In
in SASA suggests that the mutant protein underwent contrast, the C59G mutation in CCL2 leads to an increase
substantial changes in its solvent accessibility, potentially in hydrogen bond formation (total hydrogen bonds: 4195)
affecting its stability and interactions with other molecules (Figure 7B). This directly correlates with the binding
(Figure 6C and D). affinity and stability of the receptor–ligand interaction,
During the simulation, the native protein maintained which suggests that the C59G mutation enhances receptor–
a stable SASA value, indicating its stability. Conversely, ligand interactions by promoting stronger and more stable
the mutant structures exhibited higher SASA values hydrogen bonding.
with notable fluctuations, suggesting increased solvent 3.9. Effects of CCL2 mutants on the protein
accessibility and alterations in its interactions with other secondary and tertiary structures
molecules.
The I-TASSER-modeled structure of CCL2 (native
3.8. Hydrogen bonding analysis of native and and mutant) was evaluated using UCSF Chimera
mutant CCL2 proteins calculations for homology modeling. This evaluation
MD simulations were used to monitor the interactions involved comparative modeling by satisfaction of spatial
between CCR2 and CCL2. The majority of the important restraints, which requires a known related structure (i.e., a
protein–peptide interactions were hydrogen bonds (Figure 7). template) and a target-template sequence alignment. The
Volume 3 Issue 4 (2024) 15 doi: 10.36922/td.3891