Page 47 - TD-4-3
P. 47
Tumor Discovery HRD genomic alterations in Chinese NSCLC
A B
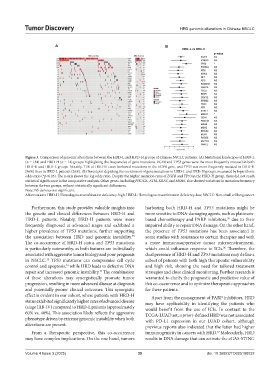
Figure 3. Comparison of genomic alterations between the HRD-L and HRD-H groups of Chinese NSCLC patients. (A) Mutational landscapes of HRD-L
(n = 134) and HRD-H (n = 14) groups highlighting the frequencies of gene mutations. EGFR and TP53 genes were the most frequently mutated in both
HRD-H and HRD-L groups. Notably, 71% of HRD-H cases harbored mutations in the EGFR gene, and TP53 was more frequently mutated in HRD-H
(86%) than in HRD-L patients (26%). (B) Forest plot depicting the enrichment of gene mutations in HRD-L and HRD-H groups, measured by logarithmic
odds ratio (*p<0.05). The x-axis shows the log odds ratio. Despite the higher mutation rates of EGFR and TP53 in the HRD-H group, these did not reach
statistical significance in the comparative analysis. Other genes, including PIK3CA, ATM, KRAS, and MSH6, also showed variations in mutation frequency
between the two groups, without statistically significant differences.
Note: NS denotes not significant.
Abbreviations: HRD-H: Homologous recombination deficiency-high; HRD-L: Homologous recombination deficiency-low; NSCLC: Non-small cell lung cancer.
Furthermore, this study provides valuable insights into harboring both HRD-H and TP53 mutations might be
the genetic and clinical differences between HRD-H and more sensitive to DNA-damaging agents, such as platinum-
35
HRD-L patients. Notably, HRD-H patients were more based chemotherapy and PARP inhibitors, due to their
frequently diagnosed at advanced stages and exhibited a impaired ability to repair DNA damage. On the other hand,
higher prevalence of TP53 mutations, further supporting the presence of TP53 mutations has been associated in
the association between HRD and genomic instability. some studies with resistance to certain therapies and with
33
The co-occurrence of HRD-H status and TP53 mutations a more immunosuppressive tumor microenvironment,
36
is particularly noteworthy, as both features are individually which could influence response to ICIs. Therefore, the
associated with aggressive tumor biology and poor prognosis dual presence of HRD-H and TP53 mutations may define a
in NSCLC. TP53 mutations can compromise cell cycle subset of patients with both high therapeutic vulnerability
32
control and apoptosis, while HRD leads to defective DNA and high risk, showing the need for tailored treatment
34
repair and increased genomic instability. The combination strategies and close clinical monitoring. Further research is
23
of these alterations may synergistically promote tumor warranted to clarify the prognostic and predictive value of
progression, resulting in more advanced disease at diagnosis this co-occurrence and to optimize therapeutic approaches
and potentially poorer clinical outcomes. This synergistic for these patients.
effect is evident in our cohort, where patients with HRD-H Apart from the management of PARP inhibitors, HRD
status exhibited significantly higher rates of advanced disease may have applicability in identifying the patients who
(stage IIIB-IV) compared to HRD-L patients (approximately would benefit from the use of ICIs. In contrast to the
60% vs. 40%). This association likely reflects the aggressive TCGA-LUAD set, a priori-defined HRD was not associated
phenotype driven by extreme genomic instability when both with PD-L1 expression in our LUAD cohort, although
alterations are present. previous reports also indicated that the latter had higher
From a therapeutic perspective, this co-occurrence immunogenicity in cancers with HRD. Molecularly, HRD
37
may have complex implications. On the one hand, tumors results in DNA damage that can activate the cGAS-STING
Volume 4 Issue 3 (2025) 39 doi: 10.36922/TD025180032