Page 80 - AIH-2-3
P. 80
Artificial Intelligence in Health CNN model for leukemia diagnosis
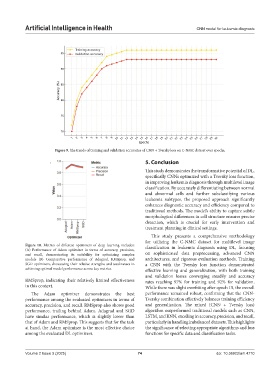
Figure 9. The trends of training and validation accuracies of CNN + Tversky loss on C-NMC dataset over epochs.
5. Conclusion
This study demonstrates the transformative potential of DL,
specifically CNNs optimized with a Tversky loss function,
in improving leukemia diagnosis through multilevel image
classification. By accurately differentiating between normal
and abnormal cells and further subclassifying various
leukemia subtypes, the proposed approach significantly
enhances diagnostic accuracy and efficiency compared to
traditional methods. The model’s ability to capture subtle
morphological differences in cell structure ensures precise
detection, which is crucial for early intervention and
treatment planning in clinical settings.
This study presents a comprehensive methodology
for utilizing the C-NMC dataset for multilevel image
Figure 10. Metrics of different optimizers of deep learning includes: classification in leukemia diagnosis using DL, focusing
(A) Performance of Adam optimizer in terms of accuracy, precision,
and recall, demonstrating its suitability for optimizing complex on sophisticated data preprocessing, advanced CNN
models (B) Comparative performance of Adagrad, RMSprop, and architectures, and rigorous evaluation methods. Training
SGD optimizers, showcasing their relative strengths and weaknesses in a CNN with the Tversky loss function demonstrated
achieving optimal model performance across key metrics. effective learning and generalization, with both training
and validation losses converging steadily and accuracy
RMSprop, indicating their relatively limited effectiveness rates reaching 97% for training and 92% for validation.
in this context. While there was slight overfitting after epoch 15, the overall
The Adam optimizer demonstrates the best performance remained robust, confirming that the CNN-
performance among the evaluated optimizers in terms of Tversky combination effectively balances training efficiency
accuracy, precision, and recall. RMSprop also shows good and generalization. The mixed (CNN + Tversky loss)
performance, trailing behind Adam. Adagrad and SGD algorithm outperformed traditional models such as CNN,
have similar performance, which is slightly lower than LSTM, and RNN, excelling in accuracy, precision, and recall,
that of Adam and RMSprop. This suggests that for the task particularly in handling imbalanced datasets. This highlights
at hand, the Adam optimizer is the most effective choice the significance of selecting appropriate algorithms and loss
among the evaluated DL optimizers. functions for specific data and classification tasks.
Volume 2 Issue 3 (2025) 74 doi: 10.36922/aih.4710