Page 54 - AN-2-4
P. 54
Advanced Neurology Genomic insights into Alzheimer
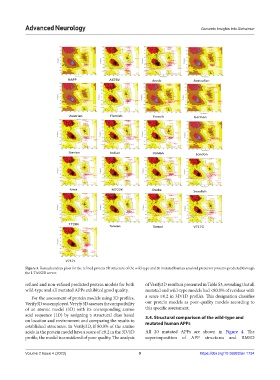
Figure 3. Ramachandran plots for the refined protein 3D structure of the wild-type and 20 mutated human amyloid precursor proteins predicted through
the I-TASSER server.
refined and non-refined predicted protein models for both of Verify3D results is presented in Table S3, revealing that all
wild-type and all mutated APPs exhibited good quality. mutated and wild-type models had <80.0% of residues with
For the assessment of protein models using 3D profiles, a score ≥0.2 in 3D/1D profiles. This designation classifies
Verify3D was employed. Veryfy 3D assesses the compatibility our protein models as poor-quality models according to
of an atomic model (3D) with its corresponding amino this specific assessment.
acid sequence (1D) by assigning a structural class based 3.4. Structural comparison of the wild-type and
on location and environment and comparing the results to mutated human APPs
established structures. In Verify3D, if 80.0% of the amino
acids in the protein model have a score of ≥0.2 in the 3D/1D All 20 mutated APPs are shown in Figure 4. The
profile, the model is considered of poor quality. The analysis superimposition of APP structures and RMSD
Volume 2 Issue 4 (2023) 9 https://doi.org/10.36922/an.1734