Page 168 - EJMO-9-2
P. 168
Eurasian Journal of
Medicine and Oncology Genetic insights into CAD drug targets
MF enrichment analysis revealed the importance of these diabetes, most of which are related to metabolic disorders,
genes in binding and enzyme activity (e.g., G-quadruplex immune responses, and chronic diseases.
DNA binding, phospholipase A1 activity). These results
illustrate the broad involvement of genes associated with 3.8. PPI network
CA in various biological functions. In addition, KEGG We loaded the candidate target genes into the STRING
enrichment suggested that candidate target genes are database to generate a PPI network and visualized the
involved in multiple disease pathways (Figure 5B). The results in Cytoscape (Figure 6A). The network contains
main enriched diseases include Alzheimer’s disease, drug 37 nodes and 107 edges, representing the complex
resistance, asthma, cholesterol metabolism, and type I interactions between genes and their corresponding
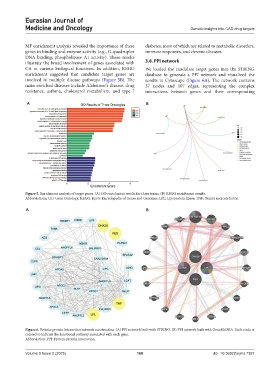
A B
Figure 5. Enrichment analysis of target genes. (A) GO enrichment results for three terms. (B) KEGG enrichment results.
Abbreviations: GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; LPL: Lipoprotein lipase; TNF: Tumor necrosis factor.
A B
Figure 6. Protein-protein interaction network construction. (A) PPI network built with STRING. (B) PPI network built with GeneMANIA. Each circle is
colored to indicate the functional pathway associated with each gene.
Abbreviation: PPI: Protein-protein interaction.
Volume 9 Issue 2 (2025) 160 doi: 10.36922/ejmo.7387